Please be patient as the page loads
|
ARFG1_HUMAN
|
||||||
| SwissProt Accessions | Q8N6T3, Q96KC4, Q96T02, Q9NSU3, Q9NVF6, Q9UIL0 | Gene names | ARFGAP1, ARF1GAP | |||
|
Domain Architecture |
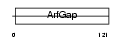
|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ARFG1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8N6T3, Q96KC4, Q96T02, Q9NSU3, Q9NVF6, Q9UIL0 | Gene names | ARFGAP1, ARF1GAP | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
ARFG1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.990657 (rank : 2) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9EPJ9, Q3TI52, Q8BMM6, Q8BMQ7 | Gene names | Arfgap1, Arf1gap | |||
|
Domain Architecture |
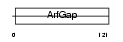
|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
ARFG3_HUMAN
|
||||||
| θ value | 5.06226e-27 (rank : 3) | NC score | 0.839931 (rank : 3) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NP61, Q9BSC6, Q9H9J0, Q9NT10, Q9NUP5, Q9Y4V3, Q9Y4V4 | Gene names | ARFGAP3, ARFGAP1 | |||
|
Domain Architecture |
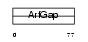
|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
ARFG3_MOUSE
|
||||||
| θ value | 3.62785e-25 (rank : 4) | NC score | 0.832707 (rank : 4) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D8S3, Q99KN8 | Gene names | Arfgap3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
DDEF2_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 5) | NC score | 0.596552 (rank : 16) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
DDEF2_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 6) | NC score | 0.594018 (rank : 17) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
CENB5_HUMAN
|
||||||
| θ value | 8.38298e-14 (rank : 7) | NC score | 0.574700 (rank : 24) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
SMP1L_HUMAN
|
||||||
| θ value | 1.09485e-13 (rank : 8) | NC score | 0.771322 (rank : 6) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WU79, Q5QPL2, Q96C93, Q9NST2, Q9UJL8 | Gene names | SMAP1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like. | |||||
|
SMP1L_MOUSE
|
||||||
| θ value | 1.09485e-13 (rank : 9) | NC score | 0.770003 (rank : 8) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TN29, Q3U798 | Gene names | Smap1l, Smap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like (Stromal membrane- associated protein 2). | |||||
|
SMAP1_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 10) | NC score | 0.771248 (rank : 7) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
DDEF1_MOUSE
|
||||||
| θ value | 3.18553e-13 (rank : 11) | NC score | 0.611570 (rank : 13) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
SMAP1_HUMAN
|
||||||
| θ value | 3.18553e-13 (rank : 12) | NC score | 0.771613 (rank : 5) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
CENB2_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 13) | NC score | 0.561088 (rank : 27) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
CENG3_MOUSE
|
||||||
| θ value | 9.26847e-13 (rank : 14) | NC score | 0.592902 (rank : 19) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8VHH5, Q812F7 | Gene names | Centg3, Agap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
CENB1_HUMAN
|
||||||
| θ value | 2.69671e-12 (rank : 15) | NC score | 0.576737 (rank : 23) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
CENG3_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 16) | NC score | 0.591552 (rank : 20) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96P47, Q59EN0, Q96RK3 | Gene names | CENTG3, AGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
CENG1_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 17) | NC score | 0.592964 (rank : 18) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
DDFL1_HUMAN
|
||||||
| θ value | 6.00763e-12 (rank : 18) | NC score | 0.639608 (rank : 12) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8TDY4, Q6P9F4, Q86UY1, Q9NXK2 | Gene names | DDEFL1, UPLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1 (Protein up- regulated in liver cancer 1). | |||||
|
CENA1_HUMAN
|
||||||
| θ value | 1.02475e-11 (rank : 19) | NC score | 0.737014 (rank : 9) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O75689 | Gene names | CENTA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 1 (Putative MAPK-activating protein PM25). | |||||
|
CENG1_MOUSE
|
||||||
| θ value | 1.02475e-11 (rank : 20) | NC score | 0.598002 (rank : 15) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
DDFL1_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 21) | NC score | 0.607396 (rank : 14) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5U464 | Gene names | Ddefl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1. | |||||
|
CENG2_MOUSE
|
||||||
| θ value | 5.08577e-11 (rank : 22) | NC score | 0.569066 (rank : 25) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BXK8, Q3UHA0, Q6ZPX9, Q8BZG0 | Gene names | Centg2, Agap1, Kiaa1099 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1). | |||||
|
CENG2_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 23) | NC score | 0.565264 (rank : 26) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UPQ3, Q541S5, Q6P9D7, Q9NV93 | Gene names | CENTG2, AGAP1, KIAA1099 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1) (GTP-binding and GTPase-activating protein 1) (GGAP1). | |||||
|
NUPL_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 24) | NC score | 0.589110 (rank : 21) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P52594, Q15277, Q4VAS0, Q4VAS1, Q4VAS3, Q53QT8, Q53R11 | Gene names | HRB, RAB, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein) (Rev- interacting protein) (Rev/Rex activation domain-binding protein). | |||||
|
NUPL_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 25) | NC score | 0.581676 (rank : 22) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K2K6, O70448, Q8BQL5, Q8CDK9 | Gene names | Hrb, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein homolog). | |||||
|
CENA2_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 26) | NC score | 0.694824 (rank : 11) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NPF8, Q8N4Q6, Q96SD5 | Gene names | CENTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-alpha 2. | |||||
|
CEND2_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 27) | NC score | 0.506234 (rank : 30) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
CENA2_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 28) | NC score | 0.701392 (rank : 10) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8R2V5 | Gene names | Centa2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 2. | |||||
|
CEND1_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 29) | NC score | 0.511110 (rank : 28) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
CEND3_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 30) | NC score | 0.494471 (rank : 33) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
CEND1_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 31) | NC score | 0.508753 (rank : 29) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
CEND3_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 32) | NC score | 0.488597 (rank : 36) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
GIT1_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 33) | NC score | 0.489767 (rank : 34) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y2X7, Q86SS0, Q9BRJ4 | Gene names | GIT1 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1) (Cool-associated and tyrosine-phosphorylated protein 1) (Cat-1). | |||||
|
GIT1_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 34) | NC score | 0.488812 (rank : 35) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q68FF6 | Gene names | Git1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1). | |||||
|
GIT2_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 35) | NC score | 0.504857 (rank : 31) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JLQ2 | Gene names | Git2 | |||
|
Domain Architecture |
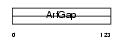
|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (Cool-interacting tyrosine-phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
GIT2_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 36) | NC score | 0.504239 (rank : 32) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14161, Q86U59, Q96CI2, Q9BV91, Q9Y5V2 | Gene names | GIT2, KIAA0148 | |||
|
Domain Architecture |
|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (GRK-interacting protein 2) (Cool-interacting tyrosine- phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
HRBL_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 37) | NC score | 0.476580 (rank : 37) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 38) | NC score | 0.069744 (rank : 59) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 39) | NC score | 0.060284 (rank : 74) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
HRBL_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 40) | NC score | 0.452651 (rank : 38) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 41) | NC score | 0.048707 (rank : 140) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DNJB6_HUMAN
|
||||||
| θ value | 0.21417 (rank : 42) | NC score | 0.029816 (rank : 145) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75190, O95806, Q9UIK6 | Gene names | DNAJB6, HSJ2, MSJ1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MSJ-1) (HHDJ1) (MRJ). | |||||
|
LIMD1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 43) | NC score | 0.020298 (rank : 151) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
DNJB6_MOUSE
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.027803 (rank : 147) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54946, Q99LA5, Q9QYI9 | Gene names | Dnajb6, Hsj2, Mrj | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MRJ) (mDj4). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.039665 (rank : 141) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.034964 (rank : 143) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
CDSN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.035218 (rank : 142) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15517, O43509, Q5SQ85, Q5STD2, Q7LA70, Q7LA71, Q86Z04, Q8IZU4, Q8IZU5, Q8IZU6, Q8N5P3, Q95IF9, Q9NP52, Q9NPE0, Q9NPG5, Q9NRH4, Q9NRH5, Q9NRH6, Q9NRH7, Q9NRH8, Q9UBH8, Q9UIN6, Q9UIN7, Q9UIN8, Q9UIN9, Q9UIP0 | Gene names | CDSN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor (S protein). | |||||
|
BCAS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.028162 (rank : 146) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75363, Q68CZ3 | Gene names | BCAS1, AIBC1, NABC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 (Novel amplified in breast cancer 1) (Amplified and overexpressed in breast cancer). | |||||
|
BRAP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.022410 (rank : 150) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z569, O43238, O75341 | Gene names | BRAP, RNF52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP) (RING finger protein 52) (NY-REN-63 antigen). | |||||
|
BRAP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.026559 (rank : 148) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99MP8, Q8CC00, Q8CHX1, Q99MP7, Q9CXX8 | Gene names | Brap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP). | |||||
|
ZCH10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.030042 (rank : 144) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.011629 (rank : 156) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
PRGR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.003589 (rank : 159) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00175 | Gene names | Pgr, Nr3c3, Pr | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.019305 (rank : 153) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.018519 (rank : 154) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
DNJB7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.020218 (rank : 152) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QYI8, Q9DA41 | Gene names | Dnajb7 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7 (mDJ5). | |||||
|
LA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.010943 (rank : 157) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P32067 | Gene names | Ssb, Ss-b | |||
|
Domain Architecture |
|
|||||
| Description | Lupus La protein homolog (La ribonucleoprotein) (La autoantigen homolog). | |||||
|
LY9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.007072 (rank : 158) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBG7, Q14775, Q5VYI3, Q9H4N5, Q9NQ24 | Gene names | LY9 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphocyte surface antigen Ly-9 precursor (Lymphocyte antigen 9) (Cell-surface molecule Ly-9) (CD229 antigen). | |||||
|
TNC6B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.023285 (rank : 149) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
XKR5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.013060 (rank : 155) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6UX68, Q5GH74 | Gene names | XKR5, XRG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 5. | |||||
|
ACBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.053861 (rank : 110) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BR61 | Gene names | ACBD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 6. | |||||
|
ACBD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.052120 (rank : 116) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D061, Q3TMN7, Q9DCU4 | Gene names | Acbd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 6. | |||||
|
AN36A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.050796 (rank : 135) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6UX02, Q86X62 | Gene names | ANKRD36, ANKRD36A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 36A. | |||||
|
ANK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.057554 (rank : 85) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
ANK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.057567 (rank : 84) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.053073 (rank : 114) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.051375 (rank : 127) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
ANKR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.055296 (rank : 102) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15327, Q96LE7 | Gene names | ANKRD1, C193, CARP, HA1A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein) (Cytokine-inducible nuclear protein) (C-193). | |||||
|
ANKR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.057091 (rank : 89) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CR42, O55014, Q3UIF7, Q3UJ39, Q792Q9 | Gene names | Ankrd1, Carp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein). | |||||
|
ANKR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.059703 (rank : 78) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9GZV1, Q5T456, Q8WUD7 | Gene names | ANKRD2, ARPP | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat domain-containing protein 2 (Skeletal muscle ankyrin repeat protein) (hArpp). | |||||
|
ANKR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.055391 (rank : 101) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WV06 | Gene names | Ankrd2, Arpp | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat domain-containing protein 2 (Skeletal muscle ankyrin repeat protein) (mArpp). | |||||
|
ANKR5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.051327 (rank : 129) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NU02, Q9H6Y9 | Gene names | ANKRD5 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 5. | |||||
|
ANKR5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.051558 (rank : 125) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D2J7 | Gene names | Ankrd5 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 5. | |||||
|
ANKR6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.063034 (rank : 69) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
ANR10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.062083 (rank : 71) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NXR5 | Gene names | ANKRD10 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
ANR10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.059194 (rank : 80) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99LW0 | Gene names | Ankrd10 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
ANR22_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.061796 (rank : 72) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5VYY1, Q8WU06 | Gene names | ANKRD22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 22. | |||||
|
ANR23_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.056256 (rank : 97) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86SG2, Q711K7, Q8NAJ7 | Gene names | ANKRD23, DARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein) (Muscle ankyrin repeat protein 3). | |||||
|
ANR23_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.053484 (rank : 111) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q812A3 | Gene names | Ankrd23, Darp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein). | |||||
|
ANR28_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.051264 (rank : 130) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15084, Q6ULS0, Q6ZT57 | Gene names | ANKRD28, KIAA0379 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR28_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.051901 (rank : 119) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q505D1 | Gene names | Ankrd28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR33_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.057007 (rank : 90) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z3H0, Q5K619, Q5K621, Q5K622, Q5K623, Q5K624, Q6ZUN0 | Gene names | ANKRD33, C12orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 33. | |||||
|
ANR35_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.054321 (rank : 107) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
ANR37_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.075481 (rank : 54) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7Z713 | Gene names | ANKRD37, LPR2BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 37 (Low-density lipoprotein receptor-related protein 2-binding protein) (hLrp2bp). | |||||
|
ANR37_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.085337 (rank : 47) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q569N2, Q8CHS0 | Gene names | Ankrd37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 37. | |||||
|
ANR39_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.073971 (rank : 56) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q53RE8, Q59FU2, Q8N5X5, Q9P0S5 | Gene names | ANKRD39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 39. | |||||
|
ANR39_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.074112 (rank : 55) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D2X0 | Gene names | Ankrd39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 39. | |||||
|
ANR41_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.082670 (rank : 50) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NAG6, Q8N8J8 | Gene names | ANKRD41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 41. | |||||
|
ANR42_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.059153 (rank : 81) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N9B4, Q49A49 | Gene names | ANKRD42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
ANR42_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.054952 (rank : 103) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3V096, Q8BX55, Q8BZT3, Q8C0T6, Q8C0X5 | Gene names | Ankrd42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
ANR44_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.053279 (rank : 113) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N8A2, Q6P480, Q86VL5, Q8IZ72, Q9UFA4 | Gene names | ANKRD44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 44. | |||||
|
ANR45_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.050007 (rank : 139) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q810N6 | Gene names | Ankrd45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
ANR46_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.083271 (rank : 49) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86W74, Q6P9B7 | Gene names | ANKRD46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 46 (Ankyrin repeat small protein) (ANK-S). | |||||
|
ANR46_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.080825 (rank : 52) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BTZ5, Q1LZK9, Q8BV97 | Gene names | Ankrd46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 46 (Ankyrin repeat small protein) (ANK-S). | |||||
|
ANR49_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.051640 (rank : 123) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WVL7, Q8NDF2, Q96JE5, Q9NXK7 | Gene names | ANKRD49, FGIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 49 (Fetal globin-inducing factor). | |||||
|
ANR50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.051643 (rank : 122) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ULJ7, Q6N064, Q6ZSE6 | Gene names | ANKRD50, KIAA1223 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 50. | |||||
|
ANR52_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.056495 (rank : 95) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NB46 | Gene names | ANKRD52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ANR52_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.056775 (rank : 93) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BTI7 | Gene names | Ankrd52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ANRA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.056800 (rank : 92) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H9E1 | Gene names | ANKRA2, ANKRA | |||
|
Domain Architecture |
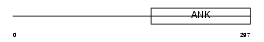
|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
ANRA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.057330 (rank : 86) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99PE2 | Gene names | Ankra2, Ankra | |||
|
Domain Architecture |
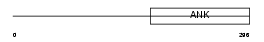
|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
ANS4B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.066948 (rank : 62) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8N8V4 | Gene names | ANKS4B, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 4B (Harmonin- interacting ankyrin repeat-containing protein) (Harp). | |||||
|
ANS4B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.073197 (rank : 57) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K3X6, Q9D8A5 | Gene names | Anks4b, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 4B (Harmonin- interacting ankyrin repeat-containing protein) (Harp). | |||||
|
ASB10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.058430 (rank : 83) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WXI3 | Gene names | ASB10 | |||
|
Domain Architecture |
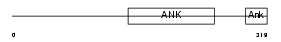
|
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
ASB10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.055865 (rank : 99) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91ZT7 | Gene names | Asb10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
ASB16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.052134 (rank : 115) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96NS5, Q8WXK0 | Gene names | ASB16 | |||
|
Domain Architecture |
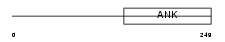
|
|||||
| Description | Ankyrin repeat and SOCS box protein 16 (ASB-16). | |||||
|
ASB16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.051096 (rank : 132) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VHS5, Q8BYT0 | Gene names | Asb16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 16 (ASB-16). | |||||
|
ASB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.050176 (rank : 138) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96Q27, Q9NSU5, Q9Y567 | Gene names | ASB2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 2 (ASB-2). | |||||
|
ASB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.051102 (rank : 131) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WV72 | Gene names | Asb3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
ASB8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.056863 (rank : 91) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H765, Q547Q2 | Gene names | ASB8 | |||
|
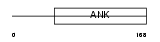
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
ASB8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.056423 (rank : 96) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91ZT9, Q8R178 | Gene names | Asb8 | |||
|
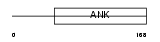
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
BARD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.054443 (rank : 105) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99728, O43574 | Gene names | BARD1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
BARD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.052101 (rank : 117) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O70445 | Gene names | Bard1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
BCL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.054417 (rank : 106) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20749 | Gene names | BCL3 | |||
|
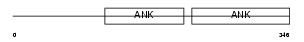
Domain Architecture |
|
|||||
| Description | B-cell lymphoma 3-encoded protein (Protein Bcl-3). | |||||
|
BCL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.054763 (rank : 104) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
CDN2D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.059585 (rank : 79) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55273, Q13102, Q6FGE9 | Gene names | CDKN2D | |||
|
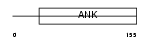
Domain Architecture |
|
|||||
| Description | Cyclin-dependent kinase 4 inhibitor D (p19-INK4d). | |||||
|
CDN2D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.050671 (rank : 136) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60773, Q60794 | Gene names | Cdkn2d | |||
|
Domain Architecture |
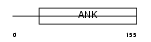
|
|||||
| Description | Cyclin-dependent kinase 4 inhibitor D (p19-INK4d). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.050349 (rank : 137) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.051608 (rank : 124) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.054118 (rank : 108) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.053318 (rank : 112) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
CT086_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.103521 (rank : 42) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BZ19, Q4VXE6 | Gene names | C20orf86 | |||
|
Domain Architecture |

|
|||||
| Description | Putative uncharacterized protein C20orf86. | |||||
|
DP13A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.106870 (rank : 40) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKG1, Q9P2B9 | Gene names | APPL1, APPL, DIP13A, KIAA1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
DP13A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.106066 (rank : 41) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
DP13B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.093307 (rank : 46) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NEU8, Q8N4R7, Q9NVL2 | Gene names | DIP13B, APPL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 beta (Dip13 beta) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 2). | |||||
|
DP13B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.102069 (rank : 43) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K3G9, Q99LT7 | Gene names | Dip13b, Appl2, Dip3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 beta (Dip13 beta) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 2). | |||||
|
EHMT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.051370 (rank : 128) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
GABP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.068161 (rank : 60) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q06547, Q06545, Q12940, Q12941, Q12942, Q8IYD0 | Gene names | GABPB2, E4TF1B, GABPB, GABPB1 | |||
|
Domain Architecture |
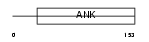
|
|||||
| Description | GA-binding protein beta chain (GABP subunit beta-2) (GABP-2) (GABP subunit beta-1) (GABPB-1) (Transcription factor E4TF1-53) (Transcription factor E4TF1-47) (Nuclear respiratory factor 2). | |||||
|
GABP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.067932 (rank : 61) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q00420, Q00421, Q8BS31, Q91YZ0, Q9QVV2 | Gene names | Gabpb2, Gabpb, Gabpb1 | |||
|
Domain Architecture |
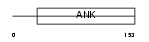
|
|||||
| Description | GA-binding protein beta chain (GABP subunit beta-2) (GABP-2) (GABP subunit beta-1) (GABPB-1). | |||||
|
GABP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.070240 (rank : 58) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P81069 | Gene names | Gabpb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GA-binding protein beta-2-1 chain (GABP subunit beta-2-1) (GABPB2-1). | |||||
|
HECD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.064975 (rank : 65) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
IKBB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.065350 (rank : 64) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15653, Q96BJ7 | Gene names | NFKBIB, IKBB, TRIP9 | |||
|
Domain Architecture |
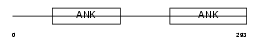
|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B) (Thyroid receptor-interacting protein 9) (TR-interacting protein 9). | |||||
|
IKBB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.075522 (rank : 53) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60778, Q9D6L5 | Gene names | Nfkbib, Ikbb | |||
|
Domain Architecture |
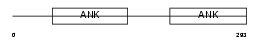
|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B). | |||||
|
INVS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.051740 (rank : 121) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
MTPN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.062486 (rank : 70) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P58546 | Gene names | MTPN | |||
|
Domain Architecture |
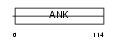
|
|||||
| Description | Myotrophin (Protein V-1). | |||||
|
MTPN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.064470 (rank : 66) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P62774, P80144, Q543M6, Q9DCN8 | Gene names | Mtpn, Gcdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotrophin (Protein V-1) (Granule cell differentiation protein). | |||||
|
MYPT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.058962 (rank : 82) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
MYPT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.059881 (rank : 77) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
MYPT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.053978 (rank : 109) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
OPHN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.100512 (rank : 44) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
OPHN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.111631 (rank : 39) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
PA2G6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.055464 (rank : 100) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60733, O75645, Q8N452, Q9UG29, Q9UIT0, Q9Y671 | Gene names | PLA2G6, IPLA2 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
PA2G6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.051918 (rank : 118) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97819, Q99LA9, Q9JK61 | Gene names | Pla2g6 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
PKHA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.084566 (rank : 48) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.082643 (rank : 51) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.057278 (rank : 88) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HB19 | Gene names | PLEKHA2, TAPP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2). | |||||
|
PKHA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.063128 (rank : 68) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ERS5, Q8BY29 | Gene names | Plekha2, Tapp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2) (PH domain-containing adaptor PHAD47). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.063265 (rank : 67) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.066633 (rank : 63) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PP16A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.051065 (rank : 133) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q923M0, Q4V9Z1 | Gene names | Ppp1r16a, Mypt3 | |||
|
Domain Architecture |
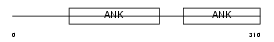
|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase targeting subunit 3). | |||||
|
PSD10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.057294 (rank : 87) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75832 | Gene names | PSMD10 | |||
|
Domain Architecture |
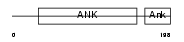
|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
PSD10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.055975 (rank : 98) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z2X2, Q9D7N8 | Gene names | Psmd10 | |||
|
Domain Architecture |
|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
RAI14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.050944 (rank : 134) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
RHG09_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.051462 (rank : 126) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
RHG26_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.094748 (rank : 45) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.051859 (rank : 120) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
TNKS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.060122 (rank : 75) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
TNKS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.056747 (rank : 94) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
USH1G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.060107 (rank : 76) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q495M9, Q8N251 | Gene names | USH1G, SANS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usher syndrome type-1G protein (Scaffold protein containing ankyrin repeats and SAM domain). | |||||
|
USH1G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.060369 (rank : 73) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80T11, Q80UG0 | Gene names | Ush1g, Sans | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usher syndrome type-1G protein homolog (Scaffold protein containing ankyrin repeats and SAM domain) (Jackson shaker protein). | |||||
|
ARFG1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8N6T3, Q96KC4, Q96T02, Q9NSU3, Q9NVF6, Q9UIL0 | Gene names | ARFGAP1, ARF1GAP | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
ARFG1_MOUSE
|
||||||
| NC score | 0.990657 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9EPJ9, Q3TI52, Q8BMM6, Q8BMQ7 | Gene names | Arfgap1, Arf1gap | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
ARFG3_HUMAN
|
||||||
| NC score | 0.839931 (rank : 3) | θ value | 5.06226e-27 (rank : 3) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NP61, Q9BSC6, Q9H9J0, Q9NT10, Q9NUP5, Q9Y4V3, Q9Y4V4 | Gene names | ARFGAP3, ARFGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
ARFG3_MOUSE
|
||||||
| NC score | 0.832707 (rank : 4) | θ value | 3.62785e-25 (rank : 4) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D8S3, Q99KN8 | Gene names | Arfgap3 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
SMAP1_HUMAN
|
||||||
| NC score | 0.771613 (rank : 5) | θ value | 3.18553e-13 (rank : 12) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SMP1L_HUMAN
|
||||||
| NC score | 0.771322 (rank : 6) | θ value | 1.09485e-13 (rank : 8) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WU79, Q5QPL2, Q96C93, Q9NST2, Q9UJL8 | Gene names | SMAP1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like. | |||||
|
SMAP1_MOUSE
|
||||||
| NC score | 0.771248 (rank : 7) | θ value | 2.43908e-13 (rank : 10) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
SMP1L_MOUSE
|
||||||
| NC score | 0.770003 (rank : 8) | θ value | 1.09485e-13 (rank : 9) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7TN29, Q3U798 | Gene names | Smap1l, Smap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like (Stromal membrane- associated protein 2). | |||||
|
CENA1_HUMAN
|
||||||
| NC score | 0.737014 (rank : 9) | θ value | 1.02475e-11 (rank : 19) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O75689 | Gene names | CENTA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 1 (Putative MAPK-activating protein PM25). | |||||
|
CENA2_MOUSE
|
||||||
| NC score | 0.701392 (rank : 10) | θ value | 1.06045e-08 (rank : 28) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8R2V5 | Gene names | Centa2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-alpha 2. | |||||
|
CENA2_HUMAN
|
||||||
| NC score | 0.694824 (rank : 11) | θ value | 3.64472e-09 (rank : 26) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NPF8, Q8N4Q6, Q96SD5 | Gene names | CENTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-alpha 2. | |||||
|
DDFL1_HUMAN
|
||||||
| NC score | 0.639608 (rank : 12) | θ value | 6.00763e-12 (rank : 18) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8TDY4, Q6P9F4, Q86UY1, Q9NXK2 | Gene names | DDEFL1, UPLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1 (Protein up- regulated in liver cancer 1). | |||||
|
DDEF1_MOUSE
|
||||||
| NC score | 0.611570 (rank : 13) | θ value | 3.18553e-13 (rank : 11) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
DDFL1_MOUSE
|
||||||
| NC score | 0.607396 (rank : 14) | θ value | 3.89403e-11 (rank : 21) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5U464 | Gene names | Ddefl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor-like 1. | |||||
|
CENG1_MOUSE
|
||||||
| NC score | 0.598002 (rank : 15) | θ value | 1.02475e-11 (rank : 20) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
DDEF2_HUMAN
|
||||||
| NC score | 0.596552 (rank : 16) | θ value | 2.20605e-14 (rank : 5) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43150 | Gene names | DDEF2, KIAA0400 | |||
|
Domain Architecture |

|
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
DDEF2_MOUSE
|
||||||
| NC score | 0.594018 (rank : 17) | θ value | 2.20605e-14 (rank : 6) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
CENG1_HUMAN
|
||||||
| NC score | 0.592964 (rank : 18) | θ value | 4.59992e-12 (rank : 17) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99490, O00578, Q548E0, Q8IWU3 | Gene names | CENTG1, AGAP2, KIAA0167 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE) (GTP-binding and GTPase-activating protein 2) (GGAP2). | |||||
|
CENG3_MOUSE
|
||||||
| NC score | 0.592902 (rank : 19) | θ value | 9.26847e-13 (rank : 14) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8VHH5, Q812F7 | Gene names | Centg3, Agap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
CENG3_HUMAN
|
||||||
| NC score | 0.591552 (rank : 20) | θ value | 3.52202e-12 (rank : 16) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96P47, Q59EN0, Q96RK3 | Gene names | CENTG3, AGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 3 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 3) (AGAP-3) (MR1-interacting protein) (MRIP-1) (CRAM-associated GTPase) (CRAG). | |||||
|
NUPL_HUMAN
|
||||||
| NC score | 0.589110 (rank : 21) | θ value | 4.30538e-10 (rank : 24) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P52594, Q15277, Q4VAS0, Q4VAS1, Q4VAS3, Q53QT8, Q53R11 | Gene names | HRB, RAB, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein) (Rev- interacting protein) (Rev/Rex activation domain-binding protein). | |||||
|
NUPL_MOUSE
|
||||||
| NC score | 0.581676 (rank : 22) | θ value | 1.25267e-09 (rank : 25) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K2K6, O70448, Q8BQL5, Q8CDK9 | Gene names | Hrb, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoporin-like protein RIP (HIV-1 Rev-binding protein homolog). | |||||
|
CENB1_HUMAN
|
||||||
| NC score | 0.576737 (rank : 23) | θ value | 2.69671e-12 (rank : 15) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15027 | Gene names | CENTB1, KIAA0050 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 1 (Cnt-b1). | |||||
|
CENB5_HUMAN
|
||||||
| NC score | 0.574700 (rank : 24) | θ value | 8.38298e-14 (rank : 7) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
CENG2_MOUSE
|
||||||
| NC score | 0.569066 (rank : 25) | θ value | 5.08577e-11 (rank : 22) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BXK8, Q3UHA0, Q6ZPX9, Q8BZG0 | Gene names | Centg2, Agap1, Kiaa1099 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1). | |||||
|
CENG2_HUMAN
|
||||||
| NC score | 0.565264 (rank : 26) | θ value | 6.64225e-11 (rank : 23) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UPQ3, Q541S5, Q6P9D7, Q9NV93 | Gene names | CENTG2, AGAP1, KIAA1099 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP-1) (GTP-binding and GTPase-activating protein 1) (GGAP1). | |||||
|
CENB2_HUMAN
|
||||||
| NC score | 0.561088 (rank : 27) | θ value | 5.43371e-13 (rank : 13) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15057, Q9UQR3 | Gene names | CENTB2, KIAA0041 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 2 (Cnt-b2). | |||||
|
CEND1_HUMAN
|
||||||
| NC score | 0.511110 (rank : 28) | θ value | 4.45536e-07 (rank : 29) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WZ64, Q4W5D2, Q7Z2L5, Q96L70, Q96P49, Q9Y4E4 | Gene names | CENTD1, ARAP2, KIAA0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 2) (PARX protein). | |||||
|
CEND1_MOUSE
|
||||||
| NC score | 0.508753 (rank : 29) | θ value | 1.69304e-06 (rank : 31) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
CEND2_HUMAN
|
||||||
| NC score | 0.506234 (rank : 30) | θ value | 6.21693e-09 (rank : 27) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96P48, O94879, Q59FI7, Q6PHS3, Q8WU51, Q96HP6, Q96L71 | Gene names | CENTD2, ARAP1, KIAA0782 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-delta 2 (Cnt-d2) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 1). | |||||
|
GIT2_MOUSE
|
||||||
| NC score | 0.504857 (rank : 31) | θ value | 8.40245e-06 (rank : 35) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JLQ2 | Gene names | Git2 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (Cool-interacting tyrosine-phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
GIT2_HUMAN
|
||||||
| NC score | 0.504239 (rank : 32) | θ value | 1.87187e-05 (rank : 36) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14161, Q86U59, Q96CI2, Q9BV91, Q9Y5V2 | Gene names | GIT2, KIAA0148 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT2 (G protein-coupled receptor kinase- interactor 2) (GRK-interacting protein 2) (Cool-interacting tyrosine- phosphorylated protein 2) (CAT2) (CAT-2). | |||||
|
CEND3_HUMAN
|
||||||
| NC score | 0.494471 (rank : 33) | θ value | 5.81887e-07 (rank : 30) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
GIT1_HUMAN
|
||||||
| NC score | 0.489767 (rank : 34) | θ value | 8.40245e-06 (rank : 33) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y2X7, Q86SS0, Q9BRJ4 | Gene names | GIT1 | |||
|
Domain Architecture |

|
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1) (Cool-associated and tyrosine-phosphorylated protein 1) (Cat-1). | |||||
|
GIT1_MOUSE
|
||||||
| NC score | 0.488812 (rank : 35) | θ value | 8.40245e-06 (rank : 34) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q68FF6 | Gene names | Git1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ARF GTPase-activating protein GIT1 (G protein-coupled receptor kinase- interactor 1) (GRK-interacting protein 1). | |||||
|
CEND3_MOUSE
|
||||||
| NC score | 0.488597 (rank : 36) | θ value | 2.88788e-06 (rank : 32) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
HRBL_HUMAN
|
||||||
| NC score | 0.476580 (rank : 37) | θ value | 0.00102713 (rank : 37) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
HRBL_MOUSE
|
||||||
| NC score | 0.452651 (rank : 38) | θ value | 0.00390308 (rank : 40) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
OPHN1_MOUSE
|
||||||
| NC score | 0.111631 (rank : 39) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99J31, Q544K7 | Gene names | Ophn1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
DP13A_HUMAN
|
||||||
| NC score | 0.106870 (rank : 40) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKG1, Q9P2B9 | Gene names | APPL1, APPL, DIP13A, KIAA1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
DP13A_MOUSE
|
||||||
| NC score | 0.106066 (rank : 41) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
CT086_HUMAN
|
||||||
| NC score | 0.103521 (rank : 42) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BZ19, Q4VXE6 | Gene names | C20orf86 | |||
|
Domain Architecture |

|
|||||
| Description | Putative uncharacterized protein C20orf86. | |||||
|
DP13B_MOUSE
|
||||||
| NC score | 0.102069 (rank : 43) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K3G9, Q99LT7 | Gene names | Dip13b, Appl2, Dip3b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 beta (Dip13 beta) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 2). | |||||
|
OPHN1_HUMAN
|
||||||
| NC score | 0.100512 (rank : 44) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60890, Q5JQ81, Q8WX47 | Gene names | OPHN1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligophrenin 1. | |||||
|
RHG26_HUMAN
|
||||||
| NC score | 0.094748 (rank : 45) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
DP13B_HUMAN
|
||||||
| NC score | 0.093307 (rank : 46) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NEU8, Q8N4R7, Q9NVL2 | Gene names | DIP13B, APPL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 beta (Dip13 beta) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 2). | |||||
|
ANR37_MOUSE
|
||||||
| NC score | 0.085337 (rank : 47) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q569N2, Q8CHS0 | Gene names | Ankrd37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 37. | |||||
|
PKHA1_HUMAN
|
||||||
| NC score | 0.084566 (rank : 48) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
ANR46_HUMAN
|
||||||
| NC score | 0.083271 (rank : 49) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86W74, Q6P9B7 | Gene names | ANKRD46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 46 (Ankyrin repeat small protein) (ANK-S). | |||||
|
ANR41_HUMAN
|
||||||
| NC score | 0.082670 (rank : 50) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NAG6, Q8N8J8 | Gene names | ANKRD41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 41. | |||||
|
PKHA1_MOUSE
|
||||||
| NC score | 0.082643 (rank : 51) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
ANR46_MOUSE
|
||||||
| NC score | 0.080825 (rank : 52) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BTZ5, Q1LZK9, Q8BV97 | Gene names | Ankrd46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 46 (Ankyrin repeat small protein) (ANK-S). | |||||
|
IKBB_MOUSE
|
||||||
| NC score | 0.075522 (rank : 53) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60778, Q9D6L5 | Gene names | Nfkbib, Ikbb | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B). | |||||
|
ANR37_HUMAN
|
||||||
| NC score | 0.075481 (rank : 54) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7Z713 | Gene names | ANKRD37, LPR2BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 37 (Low-density lipoprotein receptor-related protein 2-binding protein) (hLrp2bp). | |||||
|
ANR39_MOUSE
|
||||||
| NC score | 0.074112 (rank : 55) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D2X0 | Gene names | Ankrd39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 39. | |||||
|
ANR39_HUMAN
|
||||||
| NC score | 0.073971 (rank : 56) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q53RE8, Q59FU2, Q8N5X5, Q9P0S5 | Gene names | ANKRD39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 39. | |||||
|
ANS4B_MOUSE
|
||||||
| NC score | 0.073197 (rank : 57) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K3X6, Q9D8A5 | Gene names | Anks4b, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 4B (Harmonin- interacting ankyrin repeat-containing protein) (Harp). | |||||
|
GABP3_MOUSE
|
||||||
| NC score | 0.070240 (rank : 58) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P81069 | Gene names | Gabpb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GA-binding protein beta-2-1 chain (GABP subunit beta-2-1) (GABPB2-1). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.069744 (rank : 59) | θ value | 0.00102713 (rank : 38) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
GABP2_HUMAN
|
||||||
| NC score | 0.068161 (rank : 60) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q06547, Q06545, Q12940, Q12941, Q12942, Q8IYD0 | Gene names | GABPB2, E4TF1B, GABPB, GABPB1 | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein beta chain (GABP subunit beta-2) (GABP-2) (GABP subunit beta-1) (GABPB-1) (Transcription factor E4TF1-53) (Transcription factor E4TF1-47) (Nuclear respiratory factor 2). | |||||
|
GABP2_MOUSE
|
||||||
| NC score | 0.067932 (rank : 61) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q00420, Q00421, Q8BS31, Q91YZ0, Q9QVV2 | Gene names | Gabpb2, Gabpb, Gabpb1 | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein beta chain (GABP subunit beta-2) (GABP-2) (GABP subunit beta-1) (GABPB-1). | |||||
|
ANS4B_HUMAN
|
||||||
| NC score | 0.066948 (rank : 62) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8N8V4 | Gene names | ANKS4B, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 4B (Harmonin- interacting ankyrin repeat-containing protein) (Harp). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.066633 (rank : 63) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
IKBB_HUMAN
|
||||||
| NC score | 0.065350 (rank : 64) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15653, Q96BJ7 | Gene names | NFKBIB, IKBB, TRIP9 | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B) (Thyroid receptor-interacting protein 9) (TR-interacting protein 9). | |||||
|
HECD1_HUMAN
|
||||||
| NC score | 0.064975 (rank : 65) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
MTPN_MOUSE
|
||||||
| NC score | 0.064470 (rank : 66) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P62774, P80144, Q543M6, Q9DCN8 | Gene names | Mtpn, Gcdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotrophin (Protein V-1) (Granule cell differentiation protein). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.063265 (rank : 67) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKHA2_MOUSE
|
||||||
| NC score | 0.063128 (rank : 68) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ERS5, Q8BY29 | Gene names | Plekha2, Tapp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2) (PH domain-containing adaptor PHAD47). | |||||
|
ANKR6_HUMAN
|
||||||
| NC score | 0.063034 (rank : 69) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
MTPN_HUMAN
|
||||||
| NC score | 0.062486 (rank : 70) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P58546 | Gene names | MTPN | |||
|
Domain Architecture |

|
|||||
| Description | Myotrophin (Protein V-1). | |||||
|
ANR10_HUMAN
|
||||||
| NC score | 0.062083 (rank : 71) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NXR5 | Gene names | ANKRD10 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
ANR22_HUMAN
|
||||||
| NC score | 0.061796 (rank : 72) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5VYY1, Q8WU06 | Gene names | ANKRD22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 22. | |||||
|
USH1G_MOUSE
|
||||||
| NC score | 0.060369 (rank : 73) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80T11, Q80UG0 | Gene names | Ush1g, Sans | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usher syndrome type-1G protein homolog (Scaffold protein containing ankyrin repeats and SAM domain) (Jackson shaker protein). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.060284 (rank : 74) | θ value | 0.00175202 (rank : 39) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TNKS1_HUMAN
|
||||||
| NC score | 0.060122 (rank : 75) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
USH1G_HUMAN
|
||||||
| NC score | 0.060107 (rank : 76) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q495M9, Q8N251 | Gene names | USH1G, SANS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usher syndrome type-1G protein (Scaffold protein containing ankyrin repeats and SAM domain). | |||||
|
MYPT1_MOUSE
|
||||||
| NC score | 0.059881 (rank : 77) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
ANKR2_HUMAN
|
||||||
| NC score | 0.059703 (rank : 78) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9GZV1, Q5T456, Q8WUD7 | Gene names | ANKRD2, ARPP | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 2 (Skeletal muscle ankyrin repeat protein) (hArpp). | |||||
|
CDN2D_HUMAN
|
||||||
| NC score | 0.059585 (rank : 79) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55273, Q13102, Q6FGE9 | Gene names | CDKN2D | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 4 inhibitor D (p19-INK4d). | |||||
|
ANR10_MOUSE
|
||||||
| NC score | 0.059194 (rank : 80) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99LW0 | Gene names | Ankrd10 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
ANR42_HUMAN
|
||||||
| NC score | 0.059153 (rank : 81) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N9B4, Q49A49 | Gene names | ANKRD42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
MYPT1_HUMAN
|
||||||
| NC score | 0.058962 (rank : 82) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
ASB10_HUMAN
|
||||||
| NC score | 0.058430 (rank : 83) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WXI3 | Gene names | ASB10 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
ANK1_MOUSE
|
||||||
| NC score | 0.057567 (rank : 84) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
ANK1_HUMAN
|
||||||
| NC score | 0.057554 (rank : 85) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
ANRA2_MOUSE
|
||||||
| NC score | 0.057330 (rank : 86) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99PE2 | Gene names | Ankra2, Ankra | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
PSD10_HUMAN
|
||||||
| NC score | 0.057294 (rank : 87) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75832 | Gene names | PSMD10 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
PKHA2_HUMAN
|
||||||
| NC score | 0.057278 (rank : 88) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HB19 | Gene names | PLEKHA2, TAPP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2). | |||||
|
ANKR1_MOUSE
|
||||||
| NC score | 0.057091 (rank : 89) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CR42, O55014, Q3UIF7, Q3UJ39, Q792Q9 | Gene names | Ankrd1, Carp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein). | |||||
|
ANR33_HUMAN
|
||||||
| NC score | 0.057007 (rank : 90) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z3H0, Q5K619, Q5K621, Q5K622, Q5K623, Q5K624, Q6ZUN0 | Gene names | ANKRD33, C12orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 33. | |||||
|
ASB8_HUMAN
|
||||||
| NC score | 0.056863 (rank : 91) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H765, Q547Q2 | Gene names | ASB8 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
ANRA2_HUMAN
|
||||||
| NC score | 0.056800 (rank : 92) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H9E1 | Gene names | ANKRA2, ANKRA | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
ANR52_MOUSE
|
||||||
| NC score | 0.056775 (rank : 93) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BTI7 | Gene names | Ankrd52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
TNKS2_HUMAN
|
||||||
| NC score | 0.056747 (rank : 94) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
ANR52_HUMAN
|
||||||
| NC score | 0.056495 (rank : 95) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NB46 | Gene names | ANKRD52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ASB8_MOUSE
|
||||||
| NC score | 0.056423 (rank : 96) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91ZT9, Q8R178 | Gene names | Asb8 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
ANR23_HUMAN
|
||||||
| NC score | 0.056256 (rank : 97) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86SG2, Q711K7, Q8NAJ7 | Gene names | ANKRD23, DARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein) (Muscle ankyrin repeat protein 3). | |||||
|
PSD10_MOUSE
|
||||||
| NC score | 0.055975 (rank : 98) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z2X2, Q9D7N8 | Gene names | Psmd10 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
ASB10_MOUSE
|
||||||
| NC score | 0.055865 (rank : 99) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91ZT7 | Gene names | Asb10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
PA2G6_HUMAN
|
||||||
| NC score | 0.055464 (rank : 100) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60733, O75645, Q8N452, Q9UG29, Q9UIT0, Q9Y671 | Gene names | PLA2G6, IPLA2 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
ANKR2_MOUSE
|
||||||
| NC score | 0.055391 (rank : 101) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WV06 | Gene names | Ankrd2, Arpp | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 2 (Skeletal muscle ankyrin repeat protein) (mArpp). | |||||
|
ANKR1_HUMAN
|
||||||
| NC score | 0.055296 (rank : 102) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15327, Q96LE7 | Gene names | ANKRD1, C193, CARP, HA1A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein) (Cytokine-inducible nuclear protein) (C-193). | |||||
|
ANR42_MOUSE
|
||||||
| NC score | 0.054952 (rank : 103) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3V096, Q8BX55, Q8BZT3, Q8C0T6, Q8C0X5 | Gene names | Ankrd42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
BCL3_MOUSE
|
||||||
| NC score | 0.054763 (rank : 104) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
BARD1_HUMAN
|
||||||
| NC score | 0.054443 (rank : 105) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99728, O43574 | Gene names | BARD1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
BCL3_HUMAN
|
||||||
| NC score | 0.054417 (rank : 106) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20749 | Gene names | BCL3 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell lymphoma 3-encoded protein (Protein Bcl-3). | |||||
|
ANR35_HUMAN
|
||||||
| NC score | 0.054321 (rank : 107) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | 0.054118 (rank : 108) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
MYPT2_HUMAN
|
||||||
| NC score | 0.053978 (rank : 109) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
ACBD6_HUMAN
|
||||||
| NC score | 0.053861 (rank : 110) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BR61 | Gene names | ACBD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 6. | |||||
|
ANR23_MOUSE
|
||||||
| NC score | 0.053484 (rank : 111) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q812A3 | Gene names | Ankrd23, Darp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein). | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | 0.053318 (rank : 112) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
ANR44_HUMAN
|
||||||
| NC score | 0.053279 (rank : 113) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N8A2, Q6P480, Q86VL5, Q8IZ72, Q9UFA4 | Gene names | ANKRD44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 44. | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.053073 (rank : 114) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ASB16_HUMAN
|
||||||
| NC score | 0.052134 (rank : 115) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96NS5, Q8WXK0 | Gene names | ASB16 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 16 (ASB-16). | |||||
|
ACBD6_MOUSE
|
||||||
| NC score | 0.052120 (rank : 116) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D061, Q3TMN7, Q9DCU4 | Gene names | Acbd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 6. | |||||
|
BARD1_MOUSE
|
||||||
| NC score | 0.052101 (rank : 117) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O70445 | Gene names | Bard1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
PA2G6_MOUSE
|
||||||
| NC score | 0.051918 (rank : 118) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97819, Q99LA9, Q9JK61 | Gene names | Pla2g6 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
ANR28_MOUSE
|
||||||
| NC score | 0.051901 (rank : 119) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q505D1 | Gene names | Ankrd28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.051859 (rank : 120) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
INVS_MOUSE
|
||||||
| NC score | 0.051740 (rank : 121) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
ANR50_HUMAN
|
||||||
| NC score | 0.051643 (rank : 122) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ULJ7, Q6N064, Q6ZSE6 | Gene names | ANKRD50, KIAA1223 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 50. | |||||
|
ANR49_HUMAN
|
||||||
| NC score | 0.051640 (rank : 123) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WVL7, Q8NDF2, Q96JE5, Q9NXK7 | Gene names | ANKRD49, FGIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 49 (Fetal globin-inducing factor). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.051608 (rank : 124) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
ANKR5_MOUSE
|
||||||
| NC score | 0.051558 (rank : 125) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D2J7 | Gene names | Ankrd5 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 5. | |||||
|
RHG09_HUMAN
|
||||||
| NC score | 0.051462 (rank : 126) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.051375 (rank : 127) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
EHMT2_HUMAN
|
||||||
| NC score | 0.051370 (rank : 128) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
ANKR5_HUMAN
|
||||||
| NC score | 0.051327 (rank : 129) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NU02, Q9H6Y9 | Gene names | ANKRD5 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 5. | |||||
|
ANR28_HUMAN
|
||||||
| NC score | 0.051264 (rank : 130) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15084, Q6ULS0, Q6ZT57 | Gene names | ANKRD28, KIAA0379 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ASB3_MOUSE
|
||||||
| NC score | 0.051102 (rank : 131) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WV72 | Gene names | Asb3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
ASB16_MOUSE
|
||||||
| NC score | 0.051096 (rank : 132) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VHS5, Q8BYT0 | Gene names | Asb16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 16 (ASB-16). | |||||
|
PP16A_MOUSE
|
||||||
| NC score | 0.051065 (rank : 133) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q923M0, Q4V9Z1 | Gene names | Ppp1r16a, Mypt3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase targeting subunit 3). | |||||
|
RAI14_HUMAN
|
||||||
| NC score | 0.050944 (rank : 134) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
AN36A_HUMAN
|
||||||
| NC score | 0.050796 (rank : 135) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6UX02, Q86X62 | Gene names | ANKRD36, ANKRD36A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 36A. | |||||
|
CDN2D_MOUSE
|
||||||
| NC score | 0.050671 (rank : 136) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60773, Q60794 | Gene names | Cdkn2d | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 4 inhibitor D (p19-INK4d). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.050349 (rank : 137) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
ASB2_HUMAN
|
||||||
| NC score | 0.050176 (rank : 138) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96Q27, Q9NSU5, Q9Y567 | Gene names | ASB2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 2 (ASB-2). | |||||
|
ANR45_MOUSE
|
||||||
| NC score | 0.050007 (rank : 139) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q810N6 | Gene names | Ankrd45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.048707 (rank : 140) | θ value | 0.0330416 (rank : 41) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.039665 (rank : 141) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
CDSN_HUMAN
|
||||||
| NC score | 0.035218 (rank : 142) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15517, O43509, Q5SQ85, Q5STD2, Q7LA70, Q7LA71, Q86Z04, Q8IZU4, Q8IZU5, Q8IZU6, Q8N5P3, Q95IF9, Q9NP52, Q9NPE0, Q9NPG5, Q9NRH4, Q9NRH5, Q9NRH6, Q9NRH7, Q9NRH8, Q9UBH8, Q9UIN6, Q9UIN7, Q9UIN8, Q9UIN9, Q9UIP0 | Gene names | CDSN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor (S protein). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.034964 (rank : 143) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
ZCH10_HUMAN
|
||||||
| NC score | 0.030042 (rank : 144) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
DNJB6_HUMAN
|
||||||
| NC score | 0.029816 (rank : 145) | θ value | 0.21417 (rank : 42) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75190, O95806, Q9UIK6 | Gene names | DNAJB6, HSJ2, MSJ1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MSJ-1) (HHDJ1) (MRJ). | |||||
|
BCAS1_HUMAN
|
||||||
| NC score | 0.028162 (rank : 146) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75363, Q68CZ3 | Gene names | BCAS1, AIBC1, NABC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 (Novel amplified in breast cancer 1) (Amplified and overexpressed in breast cancer). | |||||
|
DNJB6_MOUSE
|
||||||
| NC score | 0.027803 (rank : 147) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54946, Q99LA5, Q9QYI9 | Gene names | Dnajb6, Hsj2, Mrj | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MRJ) (mDj4). | |||||
|
BRAP_MOUSE
|
||||||
| NC score | 0.026559 (rank : 148) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99MP8, Q8CC00, Q8CHX1, Q99MP7, Q9CXX8 | Gene names | Brap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP). | |||||
|
TNC6B_HUMAN
|
||||||
| NC score | 0.023285 (rank : 149) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
BRAP_HUMAN
|
||||||
| NC score | 0.022410 (rank : 150) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z569, O43238, O75341 | Gene names | BRAP, RNF52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP) (RING finger protein 52) (NY-REN-63 antigen). | |||||
|
LIMD1_MOUSE
|
||||||
| NC score | 0.020298 (rank : 151) | θ value | 0.279714 (rank : 43) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
DNJB7_MOUSE
|
||||||
| NC score | 0.020218 (rank : 152) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QYI8, Q9DA41 | Gene names | Dnajb7 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7 (mDJ5). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.019305 (rank : 153) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.018519 (rank : 154) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
XKR5_HUMAN
|
||||||
| NC score | 0.013060 (rank : 155) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6UX68, Q5GH74 | Gene names | XKR5, XRG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 5. | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.011629 (rank : 156) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
LA_MOUSE
|
||||||
| NC score | 0.010943 (rank : 157) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P32067 | Gene names | Ssb, Ss-b | |||
|
Domain Architecture |

|
|||||
| Description | Lupus La protein homolog (La ribonucleoprotein) (La autoantigen homolog). | |||||
|
LY9_HUMAN
|
||||||
| NC score | 0.007072 (rank : 158) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBG7, Q14775, Q5VYI3, Q9H4N5, Q9NQ24 | Gene names | LY9 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphocyte surface antigen Ly-9 precursor (Lymphocyte antigen 9) (Cell-surface molecule Ly-9) (CD229 antigen). | |||||
|
PRGR_MOUSE
|
||||||
| NC score | 0.003589 (rank : 159) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00175 | Gene names | Pgr, Nr3c3, Pr | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||