Please be patient as the page loads
|
DNJB6_HUMAN
|
||||||
| SwissProt Accessions | O75190, O95806, Q9UIK6 | Gene names | DNAJB6, HSJ2, MSJ1 | |||
|
Domain Architecture |
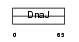
|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MSJ-1) (HHDJ1) (MRJ). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DNJB7_HUMAN
|
||||||
| θ value | 1.18549e-84 (rank : 1) | NC score | 0.992420 (rank : 4) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q7Z6W7, Q5H904, Q8WYJ7 | Gene names | DNAJB7, HSC3 | |||
|
Domain Architecture |
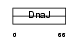
|
|||||
| Description | DnaJ homolog subfamily B member 7. | |||||
|
DNJB6_MOUSE
|
||||||
| θ value | 5.51964e-74 (rank : 2) | NC score | 0.998481 (rank : 2) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O54946, Q99LA5, Q9QYI9 | Gene names | Dnajb6, Hsj2, Mrj | |||
|
Domain Architecture |
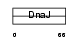
|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MRJ) (mDj4). | |||||
|
DNJB7_MOUSE
|
||||||
| θ value | 5.1662e-72 (rank : 3) | NC score | 0.992266 (rank : 5) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9QYI8, Q9DA41 | Gene names | Dnajb7 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7 (mDJ5). | |||||
|
DNJB3_MOUSE
|
||||||
| θ value | 1.11475e-66 (rank : 4) | NC score | 0.993457 (rank : 3) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O35723, Q9DAN3, Q9DAN4 | Gene names | Dnajb3, Hsj3, Msj1 | |||
|
Domain Architecture |
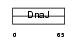
|
|||||
| Description | DnaJ homolog subfamily B member 3 (DnaJ protein homolog 3) (Heat shock J3 protein) (HSJ-3) (MSJ-1). | |||||
|
DNJB8_HUMAN
|
||||||
| θ value | 1.6097e-65 (rank : 5) | NC score | 0.991971 (rank : 7) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8NHS0 | Gene names | DNAJB8 | |||
|
Domain Architecture |
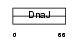
|
|||||
| Description | DnaJ homolog subfamily B member 8. | |||||
|
DNJB8_MOUSE
|
||||||
| θ value | 3.96481e-64 (rank : 6) | NC score | 0.992189 (rank : 6) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9QYI7 | Gene names | Dnajb8 | |||
|
Domain Architecture |
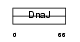
|
|||||
| Description | DnaJ homolog subfamily B member 8 (mDJ6). | |||||
|
DNJB6_HUMAN
|
||||||
| θ value | 1.91031e-50 (rank : 7) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | O75190, O95806, Q9UIK6 | Gene names | DNAJB6, HSJ2, MSJ1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MSJ-1) (HHDJ1) (MRJ). | |||||
|
DNJB2_HUMAN
|
||||||
| θ value | 2.76489e-41 (rank : 8) | NC score | 0.972478 (rank : 8) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P25686, Q8IUK1, Q8IUK2, Q96F52 | Gene names | DNAJB2, HSJ1, HSPF3 | |||
|
Domain Architecture |
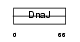
|
|||||
| Description | DnaJ homolog subfamily B member 2 (Heat shock 40 kDa protein 3) (DnaJ protein homolog 1) (HSJ-1). | |||||
|
DNJB5_HUMAN
|
||||||
| θ value | 2.51237e-26 (rank : 9) | NC score | 0.905072 (rank : 15) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O75953, Q8TDR7 | Gene names | DNAJB5, HSC40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40) (Hsp40-2). | |||||
|
DNJB5_MOUSE
|
||||||
| θ value | 2.51237e-26 (rank : 10) | NC score | 0.906168 (rank : 14) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O89114 | Gene names | Dnajb5, Hsc40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40). | |||||
|
DNJBD_MOUSE
|
||||||
| θ value | 1.37858e-24 (rank : 11) | NC score | 0.880479 (rank : 30) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q80Y75, Q8CJA2 | Gene names | Dnajb13, Tsarg, Tsarg3, Tsarg6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DNJBD_HUMAN
|
||||||
| θ value | 6.84181e-24 (rank : 12) | NC score | 0.885419 (rank : 28) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P59910, Q8IZW5 | Gene names | DNAJB13, TSARG3, TSARG6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DNJBA_MOUSE
|
||||||
| θ value | 1.16704e-23 (rank : 13) | NC score | 0.964637 (rank : 9) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9QYI5 | Gene names | Dnajb10 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 10 (mDJ8). | |||||
|
DNJB4_HUMAN
|
||||||
| θ value | 1.5242e-23 (rank : 14) | NC score | 0.900881 (rank : 20) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UDY4, Q13431 | Gene names | DNAJB4, DNAJW, HLJ1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4 (Heat shock 40 kDa protein 1 homolog) (Heat shock protein 40 homolog) (HSP40 homolog). | |||||
|
DNJB4_MOUSE
|
||||||
| θ value | 2.59989e-23 (rank : 15) | NC score | 0.901274 (rank : 18) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9D832, Q3TS92, Q9D9U2 | Gene names | Dnajb4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4. | |||||
|
DNJB1_HUMAN
|
||||||
| θ value | 1.09232e-21 (rank : 16) | NC score | 0.895623 (rank : 25) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P25685 | Gene names | DNAJB1, DNAJ1, HDJ1, HSPF1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40) (DnaJ protein homolog 1) (HDJ-1). | |||||
|
DNJB1_MOUSE
|
||||||
| θ value | 1.09232e-21 (rank : 17) | NC score | 0.895749 (rank : 24) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9QYJ3 | Gene names | Dnajb1, Hsp40, Hspf1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40). | |||||
|
DNJBC_MOUSE
|
||||||
| θ value | 2.06002e-20 (rank : 18) | NC score | 0.899821 (rank : 22) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QYI4 | Gene names | Dnajb12 | |||
|
Domain Architecture |
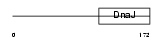
|
|||||
| Description | DnaJ homolog subfamily B member 12 (mDJ10). | |||||
|
DNJA4_MOUSE
|
||||||
| θ value | 2.69047e-20 (rank : 19) | NC score | 0.879580 (rank : 32) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9JMC3, Q543S9, Q9CTD6, Q9CUD4 | Gene names | Dnaja4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4 (MmDjA4). | |||||
|
DNJA4_HUMAN
|
||||||
| θ value | 3.51386e-20 (rank : 20) | NC score | 0.879835 (rank : 31) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8WW22, Q8N7P2 | Gene names | DNAJA4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4. | |||||
|
DNJA3_MOUSE
|
||||||
| θ value | 7.82807e-20 (rank : 21) | NC score | 0.898090 (rank : 23) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q99M87, Q8BSM0, Q99L09, Q99P71, Q99P76, Q9CT11, Q9DBJ7, Q9DC44 | Gene names | Dnaja3, Tid1 | |||
|
Domain Architecture |
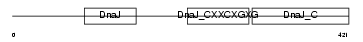
|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (mTid-1). | |||||
|
DNJA1_HUMAN
|
||||||
| θ value | 1.33526e-19 (rank : 22) | NC score | 0.865434 (rank : 34) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P31689 | Gene names | DNAJA1, DNAJ2, HDJ2, HSJ2, HSPF4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2) (HSDJ). | |||||
|
DNJA1_MOUSE
|
||||||
| θ value | 6.62687e-19 (rank : 23) | NC score | 0.862971 (rank : 35) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P63037, P54102 | Gene names | Dnaja1, Dnaj2, Hsj2, Hspf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2). | |||||
|
DNJA3_HUMAN
|
||||||
| θ value | 6.62687e-19 (rank : 24) | NC score | 0.894475 (rank : 26) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q96EY1, O75472, Q8WUJ6, Q8WXJ3, Q96D76, Q96IV1, Q9NYH8 | Gene names | DNAJA3, TID1 | |||
|
Domain Architecture |
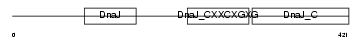
|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (hTid-1). | |||||
|
DNJBB_HUMAN
|
||||||
| θ value | 8.65492e-19 (rank : 25) | NC score | 0.902843 (rank : 16) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UBS4, Q96JC6 | Gene names | DNAJB11, EDJ, ERJ3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor (ER-associated dnaJ protein 3) (ErJ3) (ER-associated Hsp40 co-chaperone) (hDj9) (PWP1- interacting protein 4). | |||||
|
DNJBB_MOUSE
|
||||||
| θ value | 8.65492e-19 (rank : 26) | NC score | 0.902539 (rank : 17) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q99KV1, Q543I7 | Gene names | Dnajb11 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor. | |||||
|
DNJC5_HUMAN
|
||||||
| θ value | 1.92812e-18 (rank : 27) | NC score | 0.908726 (rank : 12) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9H3Z4, Q9H3Z5, Q9H7H2 | Gene names | DNAJC5, CSP | |||
|
Domain Architecture |
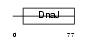
|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
DNJC5_MOUSE
|
||||||
| θ value | 1.92812e-18 (rank : 28) | NC score | 0.908571 (rank : 13) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P60904, P54101 | Gene names | Dnajc5, Csp | |||
|
Domain Architecture |
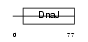
|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
DNJBC_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 29) | NC score | 0.890200 (rank : 27) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NXW2, Q9H6H0 | Gene names | DNAJB12 | |||
|
Domain Architecture |
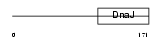
|
|||||
| Description | DnaJ homolog subfamily B member 12. | |||||
|
DNJB9_MOUSE
|
||||||
| θ value | 8.10077e-17 (rank : 30) | NC score | 0.915345 (rank : 10) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9QYI6, Q9DAW1 | Gene names | Dnajb9 | |||
|
Domain Architecture |
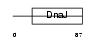
|
|||||
| Description | DnaJ homolog subfamily B member 9 (mDJ7). | |||||
|
DNJ5B_MOUSE
|
||||||
| θ value | 1.80466e-16 (rank : 31) | NC score | 0.900995 (rank : 19) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9CQ94 | Gene names | Dnajc5b | |||
|
Domain Architecture |
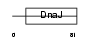
|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DCJ11_HUMAN
|
||||||
| θ value | 2.35696e-16 (rank : 32) | NC score | 0.841793 (rank : 41) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9NVH1, Q4VWF5, Q5VZN0, Q6PK20, Q6PK70, Q8NDM2, Q96CL7 | Gene names | DNAJC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DCJ11_MOUSE
|
||||||
| θ value | 2.35696e-16 (rank : 33) | NC score | 0.841914 (rank : 40) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q5U458, Q8BP83, Q8C1Z4, Q8C6U5 | Gene names | Dnajc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DNJ5B_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 34) | NC score | 0.900543 (rank : 21) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9UF47, Q969Y8 | Gene names | DNAJC5B | |||
|
Domain Architecture |
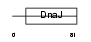
|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DJC16_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 35) | NC score | 0.845313 (rank : 39) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y2G8, Q68D57, Q86X32, Q8N5P4 | Gene names | DNAJC16, KIAA0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DNJB9_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 36) | NC score | 0.914180 (rank : 11) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UBS3 | Gene names | DNAJB9, MDG1 | |||
|
Domain Architecture |
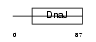
|
|||||
| Description | DnaJ homolog subfamily B member 9 (Microvascular endothelial differentiation gene 1 protein) (Mdg-1). | |||||
|
DJC16_MOUSE
|
||||||
| θ value | 1.16975e-15 (rank : 37) | NC score | 0.841755 (rank : 42) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q80TN4, Q811G1, Q8BHI2 | Gene names | Dnajc16, Kiaa0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DJC18_HUMAN
|
||||||
| θ value | 1.52774e-15 (rank : 38) | NC score | 0.873019 (rank : 33) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9H819 | Gene names | DNAJC18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DNJC7_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 39) | NC score | 0.692216 (rank : 61) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
DNJC3_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 40) | NC score | 0.734423 (rank : 54) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC3_MOUSE
|
||||||
| θ value | 4.91457e-14 (rank : 41) | NC score | 0.731551 (rank : 55) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC7_MOUSE
|
||||||
| θ value | 1.09485e-13 (rank : 42) | NC score | 0.686970 (rank : 62) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
DJC18_MOUSE
|
||||||
| θ value | 3.18553e-13 (rank : 43) | NC score | 0.849392 (rank : 38) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9CZJ9, Q8R371 | Gene names | Dnajc18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DNJA2_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 44) | NC score | 0.856843 (rank : 37) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O60884, O14711 | Gene names | DNAJA2, CPR3, HIRIP4 | |||
|
Domain Architecture |
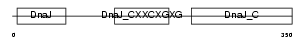
|
|||||
| Description | DnaJ homolog subfamily A member 2 (HIRA-interacting protein 4) (Cell cycle progression restoration gene 3 protein) (Dnj3) (NY-REN-14 antigen). | |||||
|
DNJA2_MOUSE
|
||||||
| θ value | 4.16044e-13 (rank : 45) | NC score | 0.857038 (rank : 36) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QYJ0 | Gene names | Dnaja2 | |||
|
Domain Architecture |
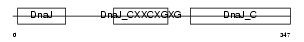
|
|||||
| Description | DnaJ homolog subfamily A member 2 (mDj3). | |||||
|
DNJ5G_HUMAN
|
||||||
| θ value | 1.2105e-12 (rank : 46) | NC score | 0.884396 (rank : 29) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8N7S2, Q8IYQ4, Q96RJ8 | Gene names | DNAJC5G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 5G (Gamma cysteine string protein) (Gamma-CSP). | |||||
|
DNJC1_MOUSE
|
||||||
| θ value | 2.0648e-12 (rank : 47) | NC score | 0.815042 (rank : 47) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q61712 | Gene names | Dnajc1, Dnajl1, Mtj1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1 (DnaJ protein homolog MTJ1). | |||||
|
DNJC1_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 48) | NC score | 0.811476 (rank : 48) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q96KC8 | Gene names | DNAJC1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1. | |||||
|
DJC17_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 49) | NC score | 0.782118 (rank : 52) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9NVM6 | Gene names | DNAJC17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DJC17_MOUSE
|
||||||
| θ value | 1.33837e-11 (rank : 50) | NC score | 0.808568 (rank : 49) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q91WT4, Q8C5R1 | Gene names | Dnajc17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DNJC9_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 51) | NC score | 0.833808 (rank : 43) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8WXX5 | Gene names | DNAJC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9 (DnaJ protein SB73). | |||||
|
DNJC9_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 52) | NC score | 0.825790 (rank : 45) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q91WN1, Q3TSG3, Q8R0E3 | Gene names | Dnajc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9. | |||||
|
DCJ14_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 53) | NC score | 0.717907 (rank : 58) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q6Y2X3, Q66K17, Q96N59, Q96T63 | Gene names | DNAJC14, DRIP78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14 (Dopamine receptor-interacting protein of 78 kDa) (DRiP78). | |||||
|
DNJC4_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 54) | NC score | 0.800819 (rank : 50) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9NNZ3, O14716 | Gene names | DNAJC4, HSPF2, MCG18 | |||
|
Domain Architecture |
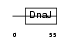
|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18) (DnaJ-like protein HSPF2). | |||||
|
WBS18_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 55) | NC score | 0.827424 (rank : 44) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P59041 | Gene names | Wbscr18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein homolog. | |||||
|
DCJ14_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 56) | NC score | 0.718764 (rank : 57) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q921R4, Q3TX73, Q8BUU3, Q9CYB7 | Gene names | Dnajc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14. | |||||
|
DJC12_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 57) | NC score | 0.715587 (rank : 59) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9R022 | Gene names | Dnajc12, Jdp1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
DNJC8_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 58) | NC score | 0.610560 (rank : 63) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75937, Q6IBA4, Q8N4Z5, Q9P051, Q9P067 | Gene names | DNAJC8, SPF31 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 8 (Splicing protein spf31). | |||||
|
DNJC8_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 59) | NC score | 0.606665 (rank : 64) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6NZB0, Q6PG72, Q8C2M6 | Gene names | Dnajc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 8. | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 60) | NC score | 0.552480 (rank : 66) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 61) | NC score | 0.552532 (rank : 65) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
DNJC4_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 62) | NC score | 0.783940 (rank : 51) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9D844, O70278 | Gene names | Dnajc4, Mcg18 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18 homolog). | |||||
|
WBS18_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 63) | NC score | 0.818876 (rank : 46) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q96LL9, Q9BSG8 | Gene names | WBSCR18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein. | |||||
|
DJC12_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 64) | NC score | 0.730161 (rank : 56) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UKB3, Q9UKB2 | Gene names | DNAJC12, JDP1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
DNJCD_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 65) | NC score | 0.529033 (rank : 68) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O75165, Q6PI82, Q6UJ77, Q6ZSW1, Q6ZUT5, Q86XG3, Q96DC1, Q9BWK9 | Gene names | DNAJC13, KIAA0678, RME8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 13 (Required for receptor-mediated endocytosis 8). | |||||
|
ZCSL3_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 66) | NC score | 0.715416 (rank : 60) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q91ZF0, Q9D9S7 | Gene names | Zcsl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3 (J-domain protein DjC7). | |||||
|
ZCSL3_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 67) | NC score | 0.754640 (rank : 53) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q6P3W2 | Gene names | ZCSL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3. | |||||
|
SACS_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 68) | NC score | 0.190155 (rank : 73) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
SACS_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 69) | NC score | 0.190831 (rank : 72) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
CU055_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 70) | NC score | 0.530079 (rank : 67) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8VCE1, Q99JX0 | Gene names | ORF28 | |||
|
Domain Architecture |
|
|||||
| Description | J domain-containing protein C21orf55 homolog. | |||||
|
ZRF1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 71) | NC score | 0.457816 (rank : 70) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
ZRF1_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 72) | NC score | 0.429083 (rank : 71) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
CU055_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 73) | NC score | 0.485848 (rank : 69) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NX36 | Gene names | C21orf55 | |||
|
Domain Architecture |
|
|||||
| Description | J domain-containing protein C21orf55. | |||||
|
ARFG1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 74) | NC score | 0.029816 (rank : 80) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N6T3, Q96KC4, Q96T02, Q9NSU3, Q9NVF6, Q9UIL0 | Gene names | ARFGAP1, ARF1GAP | |||
|
Domain Architecture |
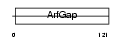
|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
AUXI_HUMAN
|
||||||
| θ value | 0.47712 (rank : 75) | NC score | 0.133236 (rank : 76) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75061, Q32M66, Q4G0K1, Q5T614, Q5T615 | Gene names | DNAJC6, KIAA0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
AUXI_MOUSE
|
||||||
| θ value | 1.06291 (rank : 76) | NC score | 0.119052 (rank : 78) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q80TZ3, Q6P2K9, Q8C7L9 | Gene names | Dnajc6, Kiaa0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
ARFG1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.024475 (rank : 82) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPJ9, Q3TI52, Q8BMM6, Q8BMQ7 | Gene names | Arfgap1, Arf1gap | |||
|
Domain Architecture |
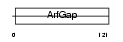
|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
GAK_HUMAN
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.025816 (rank : 81) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
GO45_MOUSE
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.010787 (rank : 85) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R2X8, Q3TJC4, Q8C0U6, Q9DAV7 | Gene names | Blzf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin 45 (Basic leucine zipper nuclear factor 1). | |||||
|
IPYR2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.021415 (rank : 83) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91VM9, Q3UPK3, Q8BTG5, Q9D1E3 | Gene names | Ppa2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inorganic pyrophosphatase 2, mitochondrial precursor (EC 3.6.1.1) (PPase 2). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.001347 (rank : 86) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
GAK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.018358 (rank : 84) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
DCJ15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.128042 (rank : 77) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y5T4, Q5T219, Q6X963 | Gene names | DNAJC15, DNAJD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 15 (Methylation-controlled J protein) (MCJ). | |||||
|
DCJ15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.097841 (rank : 79) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q78YY6 | Gene names | Dnajc15, Dnajd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 15. | |||||
|
TIM14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.134445 (rank : 74) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96DA6 | Gene names | DNAJC19, TIM14, TIMM14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial import inner membrane translocase subunit TIM14 (DnaJ homolog subfamily C member 19). | |||||
|
TIM14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.133905 (rank : 75) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9CQV7, Q8R1N1, Q9D896 | Gene names | Dnajc19, Tim14, Timm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial import inner membrane translocase subunit TIM14 (DnaJ homolog subfamily C member 19). | |||||
|
DNJB6_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.91031e-50 (rank : 7) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | O75190, O95806, Q9UIK6 | Gene names | DNAJB6, HSJ2, MSJ1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MSJ-1) (HHDJ1) (MRJ). | |||||
|
DNJB6_MOUSE
|
||||||
| NC score | 0.998481 (rank : 2) | θ value | 5.51964e-74 (rank : 2) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O54946, Q99LA5, Q9QYI9 | Gene names | Dnajb6, Hsj2, Mrj | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MRJ) (mDj4). | |||||
|
DNJB3_MOUSE
|
||||||
| NC score | 0.993457 (rank : 3) | θ value | 1.11475e-66 (rank : 4) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O35723, Q9DAN3, Q9DAN4 | Gene names | Dnajb3, Hsj3, Msj1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 3 (DnaJ protein homolog 3) (Heat shock J3 protein) (HSJ-3) (MSJ-1). | |||||
|
DNJB7_HUMAN
|
||||||
| NC score | 0.992420 (rank : 4) | θ value | 1.18549e-84 (rank : 1) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q7Z6W7, Q5H904, Q8WYJ7 | Gene names | DNAJB7, HSC3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7. | |||||
|
DNJB7_MOUSE
|
||||||
| NC score | 0.992266 (rank : 5) | θ value | 5.1662e-72 (rank : 3) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9QYI8, Q9DA41 | Gene names | Dnajb7 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7 (mDJ5). | |||||
|
DNJB8_MOUSE
|
||||||
| NC score | 0.992189 (rank : 6) | θ value | 3.96481e-64 (rank : 6) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9QYI7 | Gene names | Dnajb8 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 8 (mDJ6). | |||||
|
DNJB8_HUMAN
|
||||||
| NC score | 0.991971 (rank : 7) | θ value | 1.6097e-65 (rank : 5) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8NHS0 | Gene names | DNAJB8 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 8. | |||||
|
DNJB2_HUMAN
|
||||||
| NC score | 0.972478 (rank : 8) | θ value | 2.76489e-41 (rank : 8) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P25686, Q8IUK1, Q8IUK2, Q96F52 | Gene names | DNAJB2, HSJ1, HSPF3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 2 (Heat shock 40 kDa protein 3) (DnaJ protein homolog 1) (HSJ-1). | |||||
|
DNJBA_MOUSE
|
||||||
| NC score | 0.964637 (rank : 9) | θ value | 1.16704e-23 (rank : 13) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9QYI5 | Gene names | Dnajb10 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 10 (mDJ8). | |||||
|
DNJB9_MOUSE
|
||||||
| NC score | 0.915345 (rank : 10) | θ value | 8.10077e-17 (rank : 30) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9QYI6, Q9DAW1 | Gene names | Dnajb9 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 9 (mDJ7). | |||||
|
DNJB9_HUMAN
|
||||||
| NC score | 0.914180 (rank : 11) | θ value | 6.85773e-16 (rank : 36) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UBS3 | Gene names | DNAJB9, MDG1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 9 (Microvascular endothelial differentiation gene 1 protein) (Mdg-1). | |||||
|
DNJC5_HUMAN
|
||||||
| NC score | 0.908726 (rank : 12) | θ value | 1.92812e-18 (rank : 27) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9H3Z4, Q9H3Z5, Q9H7H2 | Gene names | DNAJC5, CSP | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
DNJC5_MOUSE
|
||||||
| NC score | 0.908571 (rank : 13) | θ value | 1.92812e-18 (rank : 28) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P60904, P54101 | Gene names | Dnajc5, Csp | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
DNJB5_MOUSE
|
||||||
| NC score | 0.906168 (rank : 14) | θ value | 2.51237e-26 (rank : 10) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O89114 | Gene names | Dnajb5, Hsc40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40). | |||||
|
DNJB5_HUMAN
|
||||||
| NC score | 0.905072 (rank : 15) | θ value | 2.51237e-26 (rank : 9) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O75953, Q8TDR7 | Gene names | DNAJB5, HSC40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40) (Hsp40-2). | |||||
|
DNJBB_HUMAN
|
||||||
| NC score | 0.902843 (rank : 16) | θ value | 8.65492e-19 (rank : 25) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UBS4, Q96JC6 | Gene names | DNAJB11, EDJ, ERJ3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor (ER-associated dnaJ protein 3) (ErJ3) (ER-associated Hsp40 co-chaperone) (hDj9) (PWP1- interacting protein 4). | |||||
|
DNJBB_MOUSE
|
||||||
| NC score | 0.902539 (rank : 17) | θ value | 8.65492e-19 (rank : 26) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q99KV1, Q543I7 | Gene names | Dnajb11 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor. | |||||
|
DNJB4_MOUSE
|
||||||
| NC score | 0.901274 (rank : 18) | θ value | 2.59989e-23 (rank : 15) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9D832, Q3TS92, Q9D9U2 | Gene names | Dnajb4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4. | |||||
|
DNJ5B_MOUSE
|
||||||
| NC score | 0.900995 (rank : 19) | θ value | 1.80466e-16 (rank : 31) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9CQ94 | Gene names | Dnajc5b | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DNJB4_HUMAN
|
||||||
| NC score | 0.900881 (rank : 20) | θ value | 1.5242e-23 (rank : 14) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UDY4, Q13431 | Gene names | DNAJB4, DNAJW, HLJ1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4 (Heat shock 40 kDa protein 1 homolog) (Heat shock protein 40 homolog) (HSP40 homolog). | |||||
|
DNJ5B_HUMAN
|
||||||
| NC score | 0.900543 (rank : 21) | θ value | 4.02038e-16 (rank : 34) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9UF47, Q969Y8 | Gene names | DNAJC5B | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DNJBC_MOUSE
|
||||||
| NC score | 0.899821 (rank : 22) | θ value | 2.06002e-20 (rank : 18) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QYI4 | Gene names | Dnajb12 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 12 (mDJ10). | |||||
|
DNJA3_MOUSE
|
||||||
| NC score | 0.898090 (rank : 23) | θ value | 7.82807e-20 (rank : 21) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q99M87, Q8BSM0, Q99L09, Q99P71, Q99P76, Q9CT11, Q9DBJ7, Q9DC44 | Gene names | Dnaja3, Tid1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (mTid-1). | |||||
|
DNJB1_MOUSE
|
||||||
| NC score | 0.895749 (rank : 24) | θ value | 1.09232e-21 (rank : 17) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9QYJ3 | Gene names | Dnajb1, Hsp40, Hspf1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40). | |||||
|
DNJB1_HUMAN
|
||||||
| NC score | 0.895623 (rank : 25) | θ value | 1.09232e-21 (rank : 16) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P25685 | Gene names | DNAJB1, DNAJ1, HDJ1, HSPF1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40) (DnaJ protein homolog 1) (HDJ-1). | |||||
|
DNJA3_HUMAN
|
||||||
| NC score | 0.894475 (rank : 26) | θ value | 6.62687e-19 (rank : 24) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q96EY1, O75472, Q8WUJ6, Q8WXJ3, Q96D76, Q96IV1, Q9NYH8 | Gene names | DNAJA3, TID1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (hTid-1). | |||||
|
DNJBC_HUMAN
|
||||||
| NC score | 0.890200 (rank : 27) | θ value | 2.7842e-17 (rank : 29) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NXW2, Q9H6H0 | Gene names | DNAJB12 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 12. | |||||
|
DNJBD_HUMAN
|
||||||
| NC score | 0.885419 (rank : 28) | θ value | 6.84181e-24 (rank : 12) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P59910, Q8IZW5 | Gene names | DNAJB13, TSARG3, TSARG6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DNJ5G_HUMAN
|
||||||
| NC score | 0.884396 (rank : 29) | θ value | 1.2105e-12 (rank : 46) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8N7S2, Q8IYQ4, Q96RJ8 | Gene names | DNAJC5G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 5G (Gamma cysteine string protein) (Gamma-CSP). | |||||
|
DNJBD_MOUSE
|
||||||
| NC score | 0.880479 (rank : 30) | θ value | 1.37858e-24 (rank : 11) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q80Y75, Q8CJA2 | Gene names | Dnajb13, Tsarg, Tsarg3, Tsarg6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DNJA4_HUMAN
|
||||||
| NC score | 0.879835 (rank : 31) | θ value | 3.51386e-20 (rank : 20) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8WW22, Q8N7P2 | Gene names | DNAJA4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4. | |||||
|
DNJA4_MOUSE
|
||||||
| NC score | 0.879580 (rank : 32) | θ value | 2.69047e-20 (rank : 19) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9JMC3, Q543S9, Q9CTD6, Q9CUD4 | Gene names | Dnaja4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4 (MmDjA4). | |||||
|
DJC18_HUMAN
|
||||||
| NC score | 0.873019 (rank : 33) | θ value | 1.52774e-15 (rank : 38) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9H819 | Gene names | DNAJC18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DNJA1_HUMAN
|
||||||
| NC score | 0.865434 (rank : 34) | θ value | 1.33526e-19 (rank : 22) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P31689 | Gene names | DNAJA1, DNAJ2, HDJ2, HSJ2, HSPF4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2) (HSDJ). | |||||
|
DNJA1_MOUSE
|
||||||
| NC score | 0.862971 (rank : 35) | θ value | 6.62687e-19 (rank : 23) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P63037, P54102 | Gene names | Dnaja1, Dnaj2, Hsj2, Hspf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2). | |||||
|
DNJA2_MOUSE
|
||||||
| NC score | 0.857038 (rank : 36) | θ value | 4.16044e-13 (rank : 45) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QYJ0 | Gene names | Dnaja2 | |||
|
Domain Architecture |
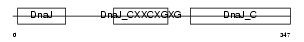
|
|||||
| Description | DnaJ homolog subfamily A member 2 (mDj3). | |||||
|
DNJA2_HUMAN
|
||||||
| NC score | 0.856843 (rank : 37) | θ value | 4.16044e-13 (rank : 44) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O60884, O14711 | Gene names | DNAJA2, CPR3, HIRIP4 | |||
|
Domain Architecture |
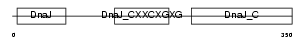
|
|||||
| Description | DnaJ homolog subfamily A member 2 (HIRA-interacting protein 4) (Cell cycle progression restoration gene 3 protein) (Dnj3) (NY-REN-14 antigen). | |||||
|
DJC18_MOUSE
|
||||||
| NC score | 0.849392 (rank : 38) | θ value | 3.18553e-13 (rank : 43) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9CZJ9, Q8R371 | Gene names | Dnajc18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DJC16_HUMAN
|
||||||
| NC score | 0.845313 (rank : 39) | θ value | 6.85773e-16 (rank : 35) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y2G8, Q68D57, Q86X32, Q8N5P4 | Gene names | DNAJC16, KIAA0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DCJ11_MOUSE
|
||||||
| NC score | 0.841914 (rank : 40) | θ value | 2.35696e-16 (rank : 33) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q5U458, Q8BP83, Q8C1Z4, Q8C6U5 | Gene names | Dnajc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DCJ11_HUMAN
|
||||||
| NC score | 0.841793 (rank : 41) | θ value | 2.35696e-16 (rank : 32) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9NVH1, Q4VWF5, Q5VZN0, Q6PK20, Q6PK70, Q8NDM2, Q96CL7 | Gene names | DNAJC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DJC16_MOUSE
|
||||||
| NC score | 0.841755 (rank : 42) | θ value | 1.16975e-15 (rank : 37) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q80TN4, Q811G1, Q8BHI2 | Gene names | Dnajc16, Kiaa0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DNJC9_HUMAN
|
||||||
| NC score | 0.833808 (rank : 43) | θ value | 1.9326e-10 (rank : 51) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8WXX5 | Gene names | DNAJC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9 (DnaJ protein SB73). | |||||
|
WBS18_MOUSE
|
||||||
| NC score | 0.827424 (rank : 44) | θ value | 7.59969e-07 (rank : 55) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P59041 | Gene names | Wbscr18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein homolog. | |||||
|
DNJC9_MOUSE
|
||||||
| NC score | 0.825790 (rank : 45) | θ value | 3.29651e-10 (rank : 52) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q91WN1, Q3TSG3, Q8R0E3 | Gene names | Dnajc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9. | |||||
|
WBS18_HUMAN
|
||||||
| NC score | 0.818876 (rank : 46) | θ value | 3.77169e-06 (rank : 63) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q96LL9, Q9BSG8 | Gene names | WBSCR18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein. | |||||
|
DNJC1_MOUSE
|
||||||
| NC score | 0.815042 (rank : 47) | θ value | 2.0648e-12 (rank : 47) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q61712 | Gene names | Dnajc1, Dnajl1, Mtj1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1 (DnaJ protein homolog MTJ1). | |||||
|
DNJC1_HUMAN
|
||||||
| NC score | 0.811476 (rank : 48) | θ value | 4.59992e-12 (rank : 48) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q96KC8 | Gene names | DNAJC1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1. | |||||
|
DJC17_MOUSE
|
||||||
| NC score | 0.808568 (rank : 49) | θ value | 1.33837e-11 (rank : 50) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q91WT4, Q8C5R1 | Gene names | Dnajc17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DNJC4_HUMAN
|
||||||
| NC score | 0.800819 (rank : 50) | θ value | 7.59969e-07 (rank : 54) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9NNZ3, O14716 | Gene names | DNAJC4, HSPF2, MCG18 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18) (DnaJ-like protein HSPF2). | |||||
|
DNJC4_MOUSE
|
||||||
| NC score | 0.783940 (rank : 51) | θ value | 3.77169e-06 (rank : 62) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9D844, O70278 | Gene names | Dnajc4, Mcg18 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18 homolog). | |||||
|
DJC17_HUMAN
|
||||||
| NC score | 0.782118 (rank : 52) | θ value | 1.33837e-11 (rank : 49) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9NVM6 | Gene names | DNAJC17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
ZCSL3_HUMAN
|
||||||
| NC score | 0.754640 (rank : 53) | θ value | 1.87187e-05 (rank : 67) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q6P3W2 | Gene names | ZCSL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3. | |||||
|
DNJC3_HUMAN
|
||||||
| NC score | 0.734423 (rank : 54) | θ value | 4.91457e-14 (rank : 40) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC3_MOUSE
|
||||||
| NC score | 0.731551 (rank : 55) | θ value | 4.91457e-14 (rank : 41) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DJC12_HUMAN
|
||||||
| NC score | 0.730161 (rank : 56) | θ value | 6.43352e-06 (rank : 64) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UKB3, Q9UKB2 | Gene names | DNAJC12, JDP1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
DCJ14_MOUSE
|
||||||
| NC score | 0.718764 (rank : 57) | θ value | 1.29631e-06 (rank : 56) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q921R4, Q3TX73, Q8BUU3, Q9CYB7 | Gene names | Dnajc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14. | |||||
|
DCJ14_HUMAN
|
||||||
| NC score | 0.717907 (rank : 58) | θ value | 1.53129e-07 (rank : 53) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q6Y2X3, Q66K17, Q96N59, Q96T63 | Gene names | DNAJC14, DRIP78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14 (Dopamine receptor-interacting protein of 78 kDa) (DRiP78). | |||||
|
DJC12_MOUSE
|
||||||
| NC score | 0.715587 (rank : 59) | θ value | 2.88788e-06 (rank : 57) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9R022 | Gene names | Dnajc12, Jdp1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
ZCSL3_MOUSE
|
||||||
| NC score | 0.715416 (rank : 60) | θ value | 1.09739e-05 (rank : 66) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q91ZF0, Q9D9S7 | Gene names | Zcsl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3 (J-domain protein DjC7). | |||||
|
DNJC7_HUMAN
|
||||||
| NC score | 0.692216 (rank : 61) | θ value | 2.20605e-14 (rank : 39) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
DNJC7_MOUSE
|
||||||
| NC score | 0.686970 (rank : 62) | θ value | 1.09485e-13 (rank : 42) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
DNJC8_HUMAN
|
||||||
| NC score | 0.610560 (rank : 63) | θ value | 2.88788e-06 (rank : 58) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75937, Q6IBA4, Q8N4Z5, Q9P051, Q9P067 | Gene names | DNAJC8, SPF31 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 8 (Splicing protein spf31). | |||||
|
DNJC8_MOUSE
|
||||||
| NC score | 0.606665 (rank : 64) | θ value | 2.88788e-06 (rank : 59) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6NZB0, Q6PG72, Q8C2M6 | Gene names | Dnajc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 8. | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.552532 (rank : 65) | θ value | 2.88788e-06 (rank : 61) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.552480 (rank : 66) | θ value | 2.88788e-06 (rank : 60) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
CU055_MOUSE
|
||||||
| NC score | 0.530079 (rank : 67) | θ value | 0.00134147 (rank : 70) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8VCE1, Q99JX0 | Gene names | ORF28 | |||
|
Domain Architecture |

|
|||||
| Description | J domain-containing protein C21orf55 homolog. | |||||
|
DNJCD_HUMAN
|
||||||
| NC score | 0.529033 (rank : 68) | θ value | 8.40245e-06 (rank : 65) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O75165, Q6PI82, Q6UJ77, Q6ZSW1, Q6ZUT5, Q86XG3, Q96DC1, Q9BWK9 | Gene names | DNAJC13, KIAA0678, RME8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 13 (Required for receptor-mediated endocytosis 8). | |||||
|
CU055_HUMAN
|
||||||
| NC score | 0.485848 (rank : 69) | θ value | 0.0330416 (rank : 73) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NX36 | Gene names | C21orf55 | |||
|
Domain Architecture |

|
|||||
| Description | J domain-containing protein C21orf55. | |||||
|
ZRF1_HUMAN
|
||||||
| NC score | 0.457816 (rank : 70) | θ value | 0.00134147 (rank : 71) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
ZRF1_MOUSE
|
||||||
| NC score | 0.429083 (rank : 71) | θ value | 0.00175202 (rank : 72) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
SACS_MOUSE
|
||||||
| NC score | 0.190831 (rank : 72) | θ value | 0.00102713 (rank : 69) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
SACS_HUMAN
|
||||||
| NC score | 0.190155 (rank : 73) | θ value | 0.00102713 (rank : 68) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZJ4, O94835 | Gene names | SACS, KIAA0730 | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
TIM14_HUMAN
|
||||||
| NC score | 0.134445 (rank : 74) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96DA6 | Gene names | DNAJC19, TIM14, TIMM14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial import inner membrane translocase subunit TIM14 (DnaJ homolog subfamily C member 19). | |||||
|
TIM14_MOUSE
|
||||||
| NC score | 0.133905 (rank : 75) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9CQV7, Q8R1N1, Q9D896 | Gene names | Dnajc19, Tim14, Timm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial import inner membrane translocase subunit TIM14 (DnaJ homolog subfamily C member 19). | |||||
|
AUXI_HUMAN
|
||||||
| NC score | 0.133236 (rank : 76) | θ value | 0.47712 (rank : 75) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75061, Q32M66, Q4G0K1, Q5T614, Q5T615 | Gene names | DNAJC6, KIAA0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
DCJ15_HUMAN
|
||||||
| NC score | 0.128042 (rank : 77) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y5T4, Q5T219, Q6X963 | Gene names | DNAJC15, DNAJD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 15 (Methylation-controlled J protein) (MCJ). | |||||
|
AUXI_MOUSE
|
||||||
| NC score | 0.119052 (rank : 78) | θ value | 1.06291 (rank : 76) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q80TZ3, Q6P2K9, Q8C7L9 | Gene names | Dnajc6, Kiaa0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
DCJ15_MOUSE
|
||||||
| NC score | 0.097841 (rank : 79) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q78YY6 | Gene names | Dnajc15, Dnajd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 15. | |||||
|
ARFG1_HUMAN
|
||||||
| NC score | 0.029816 (rank : 80) | θ value | 0.21417 (rank : 74) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N6T3, Q96KC4, Q96T02, Q9NSU3, Q9NVF6, Q9UIL0 | Gene names | ARFGAP1, ARF1GAP | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
GAK_HUMAN
|
||||||
| NC score | 0.025816 (rank : 81) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
ARFG1_MOUSE
|
||||||
| NC score | 0.024475 (rank : 82) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPJ9, Q3TI52, Q8BMM6, Q8BMQ7 | Gene names | Arfgap1, Arf1gap | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
IPYR2_MOUSE
|
||||||
| NC score | 0.021415 (rank : 83) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91VM9, Q3UPK3, Q8BTG5, Q9D1E3 | Gene names | Ppa2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inorganic pyrophosphatase 2, mitochondrial precursor (EC 3.6.1.1) (PPase 2). | |||||
|
GAK_MOUSE
|
||||||
| NC score | 0.018358 (rank : 84) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
GO45_MOUSE
|
||||||
| NC score | 0.010787 (rank : 85) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R2X8, Q3TJC4, Q8C0U6, Q9DAV7 | Gene names | Blzf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin 45 (Basic leucine zipper nuclear factor 1). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.001347 (rank : 86) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||