Please be patient as the page loads
|
IPYR2_MOUSE
|
||||||
| SwissProt Accessions | Q91VM9, Q3UPK3, Q8BTG5, Q9D1E3 | Gene names | Ppa2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inorganic pyrophosphatase 2, mitochondrial precursor (EC 3.6.1.1) (PPase 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
IPYR2_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91VM9, Q3UPK3, Q8BTG5, Q9D1E3 | Gene names | Ppa2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inorganic pyrophosphatase 2, mitochondrial precursor (EC 3.6.1.1) (PPase 2). | |||||
|
IPYR2_HUMAN
|
||||||
| θ value | 3.16907e-146 (rank : 2) | NC score | 0.996436 (rank : 2) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H2U2, Q6PG51, Q8TBW0, Q96E55, Q9H0T0, Q9NX37, Q9P033, Q9ULX0 | Gene names | PPA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inorganic pyrophosphatase 2, mitochondrial precursor (EC 3.6.1.1) (PPase 2) (Pyrophosphatase SID6-306). | |||||
|
IPYR_HUMAN
|
||||||
| θ value | 9.32813e-106 (rank : 3) | NC score | 0.986017 (rank : 3) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15181, Q5SQT7, Q6P7P4, Q9UQJ5, Q9Y5B1 | Gene names | PPA1, IOPPP, PP | |||
|
Domain Architecture |

|
|||||
| Description | Inorganic pyrophosphatase (EC 3.6.1.1) (Pyrophosphate phospho- hydrolase) (PPase). | |||||
|
IPYR_MOUSE
|
||||||
| θ value | 1.03134e-104 (rank : 4) | NC score | 0.985934 (rank : 4) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D819 | Gene names | Ppa1, Pp, Pyp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inorganic pyrophosphatase (EC 3.6.1.1) (Pyrophosphate phospho- hydrolase) (PPase). | |||||
|
DNJB2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 5) | NC score | 0.025761 (rank : 5) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P25686, Q8IUK1, Q8IUK2, Q96F52 | Gene names | DNAJB2, HSJ1, HSPF3 | |||
|
Domain Architecture |
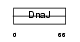
|
|||||
| Description | DnaJ homolog subfamily B member 2 (Heat shock 40 kDa protein 3) (DnaJ protein homolog 1) (HSJ-1). | |||||
|
DNJB6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 6) | NC score | 0.021415 (rank : 9) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75190, O95806, Q9UIK6 | Gene names | DNAJB6, HSJ2, MSJ1 | |||
|
Domain Architecture |
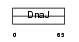
|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MSJ-1) (HHDJ1) (MRJ). | |||||
|
DNJB6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 7) | NC score | 0.021757 (rank : 7) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54946, Q99LA5, Q9QYI9 | Gene names | Dnajb6, Hsj2, Mrj | |||
|
Domain Architecture |
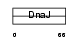
|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MRJ) (mDj4). | |||||
|
PICK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 8) | NC score | 0.018309 (rank : 10) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62083 | Gene names | Pick1, Prkcabp | |||
|
Domain Architecture |

|
|||||
| Description | PRKCA-binding protein (Protein kinase C-alpha-binding protein) (Protein interacting with C kinase 1). | |||||
|
CUL4A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 9) | NC score | 0.014601 (rank : 13) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13619, O75834, Q589T6, Q5TC62, Q6UP08, Q9UP17 | Gene names | CUL4A | |||
|
Domain Architecture |
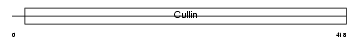
|
|||||
| Description | Cullin-4A (CUL-4A). | |||||
|
DNJB7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 10) | NC score | 0.022004 (rank : 6) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z6W7, Q5H904, Q8WYJ7 | Gene names | DNAJB7, HSC3 | |||
|
Domain Architecture |
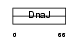
|
|||||
| Description | DnaJ homolog subfamily B member 7. | |||||
|
DNJB7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 11) | NC score | 0.021703 (rank : 8) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QYI8, Q9DA41 | Gene names | Dnajb7 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7 (mDJ5). | |||||
|
UPK1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 12) | NC score | 0.016065 (rank : 11) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75841, O60753, Q9UIM2, Q9UNX6 | Gene names | UPK1B, TSPAN20 | |||
|
Domain Architecture |

|
|||||
| Description | Uroplakin-1b (Uroplakin Ib) (UPIb) (Tetraspanin-20) (Tspan-20). | |||||
|
UPK1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 13) | NC score | 0.015597 (rank : 12) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2C6, Q8C448, Q8K4M0 | Gene names | Upk1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uroplakin-1b (Uroplakin Ib) (UPIb). | |||||
|
IPYR2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91VM9, Q3UPK3, Q8BTG5, Q9D1E3 | Gene names | Ppa2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inorganic pyrophosphatase 2, mitochondrial precursor (EC 3.6.1.1) (PPase 2). | |||||
|
IPYR2_HUMAN
|
||||||
| NC score | 0.996436 (rank : 2) | θ value | 3.16907e-146 (rank : 2) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H2U2, Q6PG51, Q8TBW0, Q96E55, Q9H0T0, Q9NX37, Q9P033, Q9ULX0 | Gene names | PPA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inorganic pyrophosphatase 2, mitochondrial precursor (EC 3.6.1.1) (PPase 2) (Pyrophosphatase SID6-306). | |||||
|
IPYR_HUMAN
|
||||||
| NC score | 0.986017 (rank : 3) | θ value | 9.32813e-106 (rank : 3) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15181, Q5SQT7, Q6P7P4, Q9UQJ5, Q9Y5B1 | Gene names | PPA1, IOPPP, PP | |||
|
Domain Architecture |

|
|||||
| Description | Inorganic pyrophosphatase (EC 3.6.1.1) (Pyrophosphate phospho- hydrolase) (PPase). | |||||
|
IPYR_MOUSE
|
||||||
| NC score | 0.985934 (rank : 4) | θ value | 1.03134e-104 (rank : 4) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D819 | Gene names | Ppa1, Pp, Pyp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inorganic pyrophosphatase (EC 3.6.1.1) (Pyrophosphate phospho- hydrolase) (PPase). | |||||
|
DNJB2_HUMAN
|
||||||
| NC score | 0.025761 (rank : 5) | θ value | 1.81305 (rank : 5) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P25686, Q8IUK1, Q8IUK2, Q96F52 | Gene names | DNAJB2, HSJ1, HSPF3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 2 (Heat shock 40 kDa protein 3) (DnaJ protein homolog 1) (HSJ-1). | |||||
|
DNJB7_HUMAN
|
||||||
| NC score | 0.022004 (rank : 6) | θ value | 6.88961 (rank : 10) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z6W7, Q5H904, Q8WYJ7 | Gene names | DNAJB7, HSC3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7. | |||||
|
DNJB6_MOUSE
|
||||||
| NC score | 0.021757 (rank : 7) | θ value | 4.03905 (rank : 7) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54946, Q99LA5, Q9QYI9 | Gene names | Dnajb6, Hsj2, Mrj | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MRJ) (mDj4). | |||||
|
DNJB7_MOUSE
|
||||||
| NC score | 0.021703 (rank : 8) | θ value | 6.88961 (rank : 11) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QYI8, Q9DA41 | Gene names | Dnajb7 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7 (mDJ5). | |||||
|
DNJB6_HUMAN
|
||||||
| NC score | 0.021415 (rank : 9) | θ value | 4.03905 (rank : 6) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75190, O95806, Q9UIK6 | Gene names | DNAJB6, HSJ2, MSJ1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MSJ-1) (HHDJ1) (MRJ). | |||||
|
PICK1_MOUSE
|
||||||
| NC score | 0.018309 (rank : 10) | θ value | 4.03905 (rank : 8) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62083 | Gene names | Pick1, Prkcabp | |||
|
Domain Architecture |

|
|||||
| Description | PRKCA-binding protein (Protein kinase C-alpha-binding protein) (Protein interacting with C kinase 1). | |||||
|
UPK1B_HUMAN
|
||||||
| NC score | 0.016065 (rank : 11) | θ value | 6.88961 (rank : 12) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75841, O60753, Q9UIM2, Q9UNX6 | Gene names | UPK1B, TSPAN20 | |||
|
Domain Architecture |

|
|||||
| Description | Uroplakin-1b (Uroplakin Ib) (UPIb) (Tetraspanin-20) (Tspan-20). | |||||
|
UPK1B_MOUSE
|
||||||
| NC score | 0.015597 (rank : 12) | θ value | 8.99809 (rank : 13) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2C6, Q8C448, Q8K4M0 | Gene names | Upk1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uroplakin-1b (Uroplakin Ib) (UPIb). | |||||
|
CUL4A_HUMAN
|
||||||
| NC score | 0.014601 (rank : 13) | θ value | 5.27518 (rank : 9) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13619, O75834, Q589T6, Q5TC62, Q6UP08, Q9UP17 | Gene names | CUL4A | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4A (CUL-4A). | |||||