Please be patient as the page loads
|
AUXI_HUMAN
|
||||||
| SwissProt Accessions | O75061, Q32M66, Q4G0K1, Q5T614, Q5T615 | Gene names | DNAJC6, KIAA0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AUXI_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | O75061, Q32M66, Q4G0K1, Q5T614, Q5T615 | Gene names | DNAJC6, KIAA0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
AUXI_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.991959 (rank : 2) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q80TZ3, Q6P2K9, Q8C7L9 | Gene names | Dnajc6, Kiaa0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
GAK_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.341935 (rank : 9) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
GAK_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.348264 (rank : 8) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
TENS1_HUMAN
|
||||||
| θ value | 6.61148e-27 (rank : 5) | NC score | 0.582311 (rank : 7) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
PTEN_HUMAN
|
||||||
| θ value | 6.84181e-24 (rank : 6) | NC score | 0.686277 (rank : 4) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60484, O00633, O02679, Q6ICT7 | Gene names | PTEN, MMAC1, TEP1 | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual- specificity protein phosphatase PTEN (EC 3.1.3.67) (EC 3.1.3.16) (EC 3.1.3.48) (Phosphatase and tensin homolog) (Mutated in multiple advanced cancers 1). | |||||
|
PTEN_MOUSE
|
||||||
| θ value | 6.84181e-24 (rank : 7) | NC score | 0.686280 (rank : 3) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08586, Q542G1 | Gene names | Pten, Mmac1 | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase PTEN (EC 3.1.3.67) (Mutated in multiple advanced cancers 1). | |||||
|
TPTE2_HUMAN
|
||||||
| θ value | 6.40375e-22 (rank : 8) | NC score | 0.674287 (rank : 6) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6XPS3, Q5VUH2, Q8WWL4, Q8WWL5 | Gene names | TPTE2, TPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase TPTE2 (EC 3.1.3.67) (TPTE and PTEN homologous inositol lipid phosphatase) (Lipid phosphatase TPIP). | |||||
|
TPTE_HUMAN
|
||||||
| θ value | 3.51386e-20 (rank : 9) | NC score | 0.674794 (rank : 5) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P56180, Q6XPS4, Q6XPS5, Q71JA8, Q8NCS8 | Gene names | TPTE | |||
|
Domain Architecture |

|
|||||
| Description | Putative tyrosine-protein phosphatase TPTE (EC 3.1.3.48) (Transmembrane phosphatase with tensin homology) (Protein BJ-HCC-5). | |||||
|
DNJ5B_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 10) | NC score | 0.169781 (rank : 10) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9CQ94 | Gene names | Dnajc5b | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 11) | NC score | 0.080276 (rank : 68) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
DNJ5B_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 12) | NC score | 0.163912 (rank : 11) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UF47, Q969Y8 | Gene names | DNAJC5B | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DNJB7_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 13) | NC score | 0.147298 (rank : 15) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7Z6W7, Q5H904, Q8WYJ7 | Gene names | DNAJB7, HSC3 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 7. | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 14) | NC score | 0.034901 (rank : 96) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
DNJB7_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 15) | NC score | 0.147339 (rank : 14) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QYI8, Q9DA41 | Gene names | Dnajb7 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7 (mDJ5). | |||||
|
DNJC5_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 16) | NC score | 0.159011 (rank : 12) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9H3Z4, Q9H3Z5, Q9H7H2 | Gene names | DNAJC5, CSP | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
DNJC5_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 17) | NC score | 0.158995 (rank : 13) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P60904, P54101 | Gene names | Dnajc5, Csp | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 18) | NC score | 0.011075 (rank : 131) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
DNJB8_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 19) | NC score | 0.144030 (rank : 16) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QYI7 | Gene names | Dnajb8 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 8 (mDJ6). | |||||
|
DNJB5_MOUSE
|
||||||
| θ value | 0.125558 (rank : 20) | NC score | 0.126529 (rank : 21) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O89114 | Gene names | Dnajb5, Hsc40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40). | |||||
|
NU153_HUMAN
|
||||||
| θ value | 0.125558 (rank : 21) | NC score | 0.072266 (rank : 77) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
DNJB5_HUMAN
|
||||||
| θ value | 0.163984 (rank : 22) | NC score | 0.123309 (rank : 25) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75953, Q8TDR7 | Gene names | DNAJB5, HSC40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40) (Hsp40-2). | |||||
|
DNJBB_MOUSE
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.124713 (rank : 22) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99KV1, Q543I7 | Gene names | Dnajb11 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor. | |||||
|
PDXL2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.061157 (rank : 84) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
DCBD1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.050273 (rank : 91) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D4J3, Q8R327, Q9D696 | Gene names | Dcbld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
CAD11_HUMAN
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.011940 (rank : 129) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55287, Q15065, Q15066, Q9UQ93, Q9UQ94 | Gene names | CDH11 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-11 precursor (Osteoblast-cadherin) (OB-cadherin) (OSF-4). | |||||
|
CAD11_MOUSE
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.011937 (rank : 130) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55288 | Gene names | Cdh11, Cad-11 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-11 precursor (Osteoblast-cadherin) (OB-cadherin) (OSF-4). | |||||
|
DNJB8_HUMAN
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.136809 (rank : 17) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8NHS0 | Gene names | DNAJB8 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 8. | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.014490 (rank : 120) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
DNJBB_HUMAN
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.119266 (rank : 30) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UBS4, Q96JC6 | Gene names | DNAJB11, EDJ, ERJ3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor (ER-associated dnaJ protein 3) (ErJ3) (ER-associated Hsp40 co-chaperone) (hDj9) (PWP1- interacting protein 4). | |||||
|
PAX9_HUMAN
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.027646 (rank : 101) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55771, Q99582, Q9UQR4 | Gene names | PAX9 | |||
|
Domain Architecture |
|
|||||
| Description | Paired box protein Pax-9. | |||||
|
PAX9_MOUSE
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.027376 (rank : 102) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47242 | Gene names | Pax9, Pax-9 | |||
|
Domain Architecture |
|
|||||
| Description | Paired box protein Pax-9. | |||||
|
TTLL5_MOUSE
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.039842 (rank : 94) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
CTND2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.032042 (rank : 98) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
DNJB4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.121070 (rank : 28) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9D832, Q3TS92, Q9D9U2 | Gene names | Dnajb4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4. | |||||
|
DNJB6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.133236 (rank : 19) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75190, O95806, Q9UIK6 | Gene names | DNAJB6, HSJ2, MSJ1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MSJ-1) (HHDJ1) (MRJ). | |||||
|
DNJB6_MOUSE
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.133283 (rank : 18) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O54946, Q99LA5, Q9QYI9 | Gene names | Dnajb6, Hsj2, Mrj | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MRJ) (mDj4). | |||||
|
DNJC7_HUMAN
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.095094 (rank : 45) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
DNJC7_MOUSE
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.094868 (rank : 47) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
CTND2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.026801 (rank : 103) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
DNJA4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.121135 (rank : 27) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WW22, Q8N7P2 | Gene names | DNAJA4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4. | |||||
|
DNJA4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.120850 (rank : 29) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JMC3, Q543S9, Q9CTD6, Q9CUD4 | Gene names | Dnaja4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4 (MmDjA4). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.028007 (rank : 100) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
DNJA1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.111691 (rank : 33) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P31689 | Gene names | DNAJA1, DNAJ2, HDJ2, HSJ2, HSPF4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2) (HSDJ). | |||||
|
DNJA1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.111690 (rank : 34) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P63037, P54102 | Gene names | Dnaja1, Dnaj2, Hsj2, Hspf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2). | |||||
|
TRIP6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.016805 (rank : 115) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15654, O15170, O15275, Q9BTB2, Q9UNT4 | Gene names | TRIP6, OIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 6 (TRIP6) (OPA-interacting protein 1) (Zyxin-related protein 1) (ZRP-1). | |||||
|
DNJB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.128751 (rank : 20) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P25685 | Gene names | DNAJB1, DNAJ1, HDJ1, HSPF1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40) (DnaJ protein homolog 1) (HDJ-1). | |||||
|
FOXI1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.014600 (rank : 119) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12951, Q14518, Q66SR7, Q8N6L8 | Gene names | FOXI1, FKHL10, FREAC6 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein I1 (Forkhead-related protein FKHL10) (Forkhead- related transcription factor 6) (FREAC-6) (Hepatocyte nuclear factor 3 forkhead homolog 3) (HNF-3/fork-head homolog 3) (HFH-3). | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.020272 (rank : 112) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
TNR6A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.043987 (rank : 93) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
ATS14_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.013112 (rank : 127) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WXS8, Q8TE55, Q8TEY8 | Gene names | ADAMTS14 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-14 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 14) (ADAM-TS 14) (ADAM-TS14). | |||||
|
LPIN3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.017110 (rank : 114) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQK8, Q5TDB9, Q9NPY8, Q9UJE5 | Gene names | LPIN3, LIPN3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-3 (Lipin 3-like). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.030814 (rank : 99) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.006728 (rank : 135) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
COE3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.023251 (rank : 105) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H4W6, Q5T6H9, Q9H4W5 | Gene names | EBF3, COE3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
DNJBA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.121317 (rank : 26) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QYI5 | Gene names | Dnajb10 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 10 (mDJ8). | |||||
|
HRBL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.036369 (rank : 95) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
COE3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.021870 (rank : 109) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08791, O08793 | Gene names | Ebf3, Coe3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
DNJB1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.123391 (rank : 24) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QYJ3 | Gene names | Dnajb1, Hsp40, Hspf1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40). | |||||
|
DNJB4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.112532 (rank : 32) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UDY4, Q13431 | Gene names | DNAJB4, DNAJW, HLJ1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4 (Heat shock 40 kDa protein 1 homolog) (Heat shock protein 40 homolog) (HSP40 homolog). | |||||
|
DNJC8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.098737 (rank : 40) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75937, Q6IBA4, Q8N4Z5, Q9P051, Q9P067 | Gene names | DNAJC8, SPF31 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 8 (Splicing protein spf31). | |||||
|
DNJC8_MOUSE
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.098079 (rank : 41) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6NZB0, Q6PG72, Q8C2M6 | Gene names | Dnajc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 8. | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.009781 (rank : 132) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
DNJB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.123633 (rank : 23) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P25686, Q8IUK1, Q8IUK2, Q96F52 | Gene names | DNAJB2, HSJ1, HSPF3 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 2 (Heat shock 40 kDa protein 3) (DnaJ protein homolog 1) (HSJ-1). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.046993 (rank : 92) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
T22D4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.019067 (rank : 113) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
DCJ14_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.096199 (rank : 43) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6Y2X3, Q66K17, Q96N59, Q96T63 | Gene names | DNAJC14, DRIP78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14 (Dopamine receptor-interacting protein of 78 kDa) (DRiP78). | |||||
|
DCJ14_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.091475 (rank : 52) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q921R4, Q3TX73, Q8BUU3, Q9CYB7 | Gene names | Dnajc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14. | |||||
|
DJC17_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.109277 (rank : 37) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NVM6 | Gene names | DNAJC17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DJC17_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.113074 (rank : 31) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q91WT4, Q8C5R1 | Gene names | Dnajc17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
INT1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.034152 (rank : 97) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P4S8, Q80UQ7, Q91Z01, Q9CTF7 | Gene names | Ints1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 1 (Int1). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.014353 (rank : 121) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
ZCSL3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.094956 (rank : 46) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6P3W2 | Gene names | ZCSL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3. | |||||
|
CO039_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.013251 (rank : 126) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
DAXX_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.013643 (rank : 125) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35613, Q9QWT8, Q9QWV3 | Gene names | Daxx | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx). | |||||
|
INT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.021202 (rank : 111) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N201, Q6NT70, Q6UX74, Q8WV40, Q96D36, Q9NTD1, Q9P2A8, Q9Y3W8 | Gene names | INTS1, KIAA1440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 1 (Int1). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.008498 (rank : 133) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
TACC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.014036 (rank : 123) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
TTLL5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.023101 (rank : 106) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6EMB2, Q9P1V5, Q9UPZ4 | Gene names | TTLL5, KIAA0998, STAMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5 (SRC1 and TIF2-associated modulatory protein). | |||||
|
BNC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.006961 (rank : 134) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01954, Q15840 | Gene names | BNC1, BNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-1. | |||||
|
CT151_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.015717 (rank : 118) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NC74, Q8N4Z9, Q9BR75, Q9H0Y9 | Gene names | C20orf151 | |||
|
Domain Architecture |
|
|||||
| Description | Uncharacterized protein C20orf151. | |||||
|
GATA5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.025884 (rank : 104) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97489 | Gene names | Gata5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
GRIP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.005035 (rank : 138) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
JIP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.022923 (rank : 107) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
JIP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.012931 (rank : 128) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
ZCSL3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.087743 (rank : 56) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q91ZF0, Q9D9S7 | Gene names | Zcsl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3 (J-domain protein DjC7). | |||||
|
ZRF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.072552 (rank : 76) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
ZRF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.071284 (rank : 81) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
AATK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.013732 (rank : 124) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.014066 (rank : 122) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
CAD15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.006662 (rank : 136) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55291 | Gene names | CDH15, CDH14, CDH3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-15 precursor (Muscle-cadherin) (M-cadherin) (Cadherin-14). | |||||
|
DCJ11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.110012 (rank : 35) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NVH1, Q4VWF5, Q5VZN0, Q6PK20, Q6PK70, Q8NDM2, Q96CL7 | Gene names | DNAJC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DCJ11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.109986 (rank : 36) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5U458, Q8BP83, Q8C1Z4, Q8C6U5 | Gene names | Dnajc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.016625 (rank : 117) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
DNJB3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.108048 (rank : 38) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O35723, Q9DAN3, Q9DAN4 | Gene names | Dnajb3, Hsj3, Msj1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily B member 3 (DnaJ protein homolog 3) (Heat shock J3 protein) (HSJ-3) (MSJ-1). | |||||
|
HRBL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.022039 (rank : 108) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
MGRN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.021651 (rank : 110) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60291, Q86W76 | Gene names | MGRN1, KIAA0544, RNF156 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase MGRN1 (EC 6.3.2.-) (Mahogunin ring finger protein 1) (RING finger protein 156). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.016668 (rank : 116) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
STAT6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.005767 (rank : 137) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52633 | Gene names | Stat6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and transcription activator 6. | |||||
|
AAK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.074820 (rank : 74) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1077 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q2M2I8, Q4ZFZ3, Q53RX6, Q9UPV4 | Gene names | AAK1, KIAA1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
AAK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.075006 (rank : 72) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
BMP2K_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.072224 (rank : 79) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NSY1, O94791, Q8IYF2, Q8N2G7, Q8NHG9, Q9NTG8 | Gene names | BMP2K, BIKE | |||
|
Domain Architecture |
|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
BMP2K_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.073136 (rank : 75) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91Z96, Q8C8L7 | Gene names | Bmp2k, Bike | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
CU055_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.052172 (rank : 89) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VCE1, Q99JX0 | Gene names | ORF28 | |||
|
Domain Architecture |
|
|||||
| Description | J domain-containing protein C21orf55 homolog. | |||||
|
DJC12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.082631 (rank : 62) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UKB3, Q9UKB2 | Gene names | DNAJC12, JDP1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
DJC12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.074862 (rank : 73) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9R022 | Gene names | Dnajc12, Jdp1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
DJC16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.080770 (rank : 66) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y2G8, Q68D57, Q86X32, Q8N5P4 | Gene names | DNAJC16, KIAA0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DJC16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.079560 (rank : 70) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80TN4, Q811G1, Q8BHI2 | Gene names | Dnajc16, Kiaa0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DJC18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.085200 (rank : 60) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H819 | Gene names | DNAJC18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DJC18_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.081553 (rank : 63) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9CZJ9, Q8R371 | Gene names | Dnajc18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DNJ5G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.106678 (rank : 39) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8N7S2, Q8IYQ4, Q96RJ8 | Gene names | DNAJC5G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 5G (Gamma cysteine string protein) (Gamma-CSP). | |||||
|
DNJA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.094744 (rank : 48) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O60884, O14711 | Gene names | DNAJA2, CPR3, HIRIP4 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily A member 2 (HIRA-interacting protein 4) (Cell cycle progression restoration gene 3 protein) (Dnj3) (NY-REN-14 antigen). | |||||
|
DNJA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.094676 (rank : 49) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QYJ0 | Gene names | Dnaja2 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily A member 2 (mDj3). | |||||
|
DNJA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.088919 (rank : 54) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96EY1, O75472, Q8WUJ6, Q8WXJ3, Q96D76, Q96IV1, Q9NYH8 | Gene names | DNAJA3, TID1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (hTid-1). | |||||
|
DNJA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.089423 (rank : 53) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99M87, Q8BSM0, Q99L09, Q99P71, Q99P76, Q9CT11, Q9DBJ7, Q9DC44 | Gene names | Dnaja3, Tid1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (mTid-1). | |||||
|
DNJB9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.091799 (rank : 50) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UBS3 | Gene names | DNAJB9, MDG1 | |||
|
Domain Architecture |
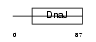
|
|||||
| Description | DnaJ homolog subfamily B member 9 (Microvascular endothelial differentiation gene 1 protein) (Mdg-1). | |||||
|
DNJB9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.091689 (rank : 51) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QYI6, Q9DAW1 | Gene names | Dnajb9 | |||
|
Domain Architecture |
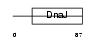
|
|||||
| Description | DnaJ homolog subfamily B member 9 (mDJ7). | |||||
|
DNJBC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.086675 (rank : 58) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NXW2, Q9H6H0 | Gene names | DNAJB12 | |||
|
Domain Architecture |
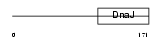
|
|||||
| Description | DnaJ homolog subfamily B member 12. | |||||
|
DNJBC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.088472 (rank : 55) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QYI4 | Gene names | Dnajb12 | |||
|
Domain Architecture |
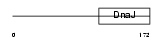
|
|||||
| Description | DnaJ homolog subfamily B member 12 (mDJ10). | |||||
|
DNJBD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.096373 (rank : 42) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P59910, Q8IZW5 | Gene names | DNAJB13, TSARG3, TSARG6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DNJBD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.095503 (rank : 44) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q80Y75, Q8CJA2 | Gene names | Dnajb13, Tsarg, Tsarg3, Tsarg6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DNJC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.079759 (rank : 69) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96KC8 | Gene names | DNAJC1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1. | |||||
|
DNJC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.080349 (rank : 67) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61712 | Gene names | Dnajc1, Dnajl1, Mtj1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1 (DnaJ protein homolog MTJ1). | |||||
|
DNJC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.072253 (rank : 78) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.071930 (rank : 80) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
DNJC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.081232 (rank : 64) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NNZ3, O14716 | Gene names | DNAJC4, HSPF2, MCG18 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18) (DnaJ-like protein HSPF2). | |||||
|
DNJC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.079125 (rank : 71) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D844, O70278 | Gene names | Dnajc4, Mcg18 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18 homolog). | |||||
|
DNJC9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.083384 (rank : 61) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WXX5 | Gene names | DNAJC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9 (DnaJ protein SB73). | |||||
|
DNJC9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.080810 (rank : 65) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91WN1, Q3TSG3, Q8R0E3 | Gene names | Dnajc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9. | |||||
|
DNJCD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.053262 (rank : 88) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75165, Q6PI82, Q6UJ77, Q6ZSW1, Q6ZUT5, Q86XG3, Q96DC1, Q9BWK9 | Gene names | DNAJC13, KIAA0678, RME8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 13 (Required for receptor-mediated endocytosis 8). | |||||
|
K2002_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.057238 (rank : 87) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H792, Q6ZS78, Q8NCM3 | Gene names | KIAA2002 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized tyrosine-protein kinase KIAA2002 (EC 2.7.10.2). | |||||
|
PRP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.050796 (rank : 90) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
SEC63_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.058314 (rank : 85) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SEC63_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.058059 (rank : 86) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
TENS4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.068548 (rank : 83) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IZW8, Q71RB7, Q8WV64, Q96JV4 | Gene names | TNS4, CTEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor (C-terminal tensin-like protein). | |||||
|
TENS4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.069771 (rank : 82) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BZ33, Q3TCM8, Q7TNR5 | Gene names | Tns4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor. | |||||
|
WBS18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.085463 (rank : 59) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96LL9, Q9BSG8 | Gene names | WBSCR18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein. | |||||
|
WBS18_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.087259 (rank : 57) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P59041 | Gene names | Wbscr18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein homolog. | |||||
|
AUXI_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | O75061, Q32M66, Q4G0K1, Q5T614, Q5T615 | Gene names | DNAJC6, KIAA0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
AUXI_MOUSE
|
||||||
| NC score | 0.991959 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q80TZ3, Q6P2K9, Q8C7L9 | Gene names | Dnajc6, Kiaa0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
PTEN_MOUSE
|
||||||
| NC score | 0.686280 (rank : 3) | θ value | 6.84181e-24 (rank : 7) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08586, Q542G1 | Gene names | Pten, Mmac1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase PTEN (EC 3.1.3.67) (Mutated in multiple advanced cancers 1). | |||||
|
PTEN_HUMAN
|
||||||
| NC score | 0.686277 (rank : 4) | θ value | 6.84181e-24 (rank : 6) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60484, O00633, O02679, Q6ICT7 | Gene names | PTEN, MMAC1, TEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual- specificity protein phosphatase PTEN (EC 3.1.3.67) (EC 3.1.3.16) (EC 3.1.3.48) (Phosphatase and tensin homolog) (Mutated in multiple advanced cancers 1). | |||||
|
TPTE_HUMAN
|
||||||
| NC score | 0.674794 (rank : 5) | θ value | 3.51386e-20 (rank : 9) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P56180, Q6XPS4, Q6XPS5, Q71JA8, Q8NCS8 | Gene names | TPTE | |||
|
Domain Architecture |

|
|||||
| Description | Putative tyrosine-protein phosphatase TPTE (EC 3.1.3.48) (Transmembrane phosphatase with tensin homology) (Protein BJ-HCC-5). | |||||
|
TPTE2_HUMAN
|
||||||
| NC score | 0.674287 (rank : 6) | θ value | 6.40375e-22 (rank : 8) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6XPS3, Q5VUH2, Q8WWL4, Q8WWL5 | Gene names | TPTE2, TPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase TPTE2 (EC 3.1.3.67) (TPTE and PTEN homologous inositol lipid phosphatase) (Lipid phosphatase TPIP). | |||||
|
TENS1_HUMAN
|
||||||
| NC score | 0.582311 (rank : 7) | θ value | 6.61148e-27 (rank : 5) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
GAK_MOUSE
|
||||||
| NC score | 0.348264 (rank : 8) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
GAK_HUMAN
|
||||||
| NC score | 0.341935 (rank : 9) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
DNJ5B_MOUSE
|
||||||
| NC score | 0.169781 (rank : 10) | θ value | 0.0193708 (rank : 10) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9CQ94 | Gene names | Dnajc5b | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DNJ5B_HUMAN
|
||||||
| NC score | 0.163912 (rank : 11) | θ value | 0.0252991 (rank : 12) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UF47, Q969Y8 | Gene names | DNAJC5B | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5B (Beta cysteine string protein) (Beta-CSP). | |||||
|
DNJC5_HUMAN
|
||||||
| NC score | 0.159011 (rank : 12) | θ value | 0.0736092 (rank : 16) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9H3Z4, Q9H3Z5, Q9H7H2 | Gene names | DNAJC5, CSP | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
DNJC5_MOUSE
|
||||||
| NC score | 0.158995 (rank : 13) | θ value | 0.0736092 (rank : 17) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P60904, P54101 | Gene names | Dnajc5, Csp | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 5 (Cysteine string protein) (CSP). | |||||
|
DNJB7_MOUSE
|
||||||
| NC score | 0.147339 (rank : 14) | θ value | 0.0736092 (rank : 15) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QYI8, Q9DA41 | Gene names | Dnajb7 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7 (mDJ5). | |||||
|
DNJB7_HUMAN
|
||||||
| NC score | 0.147298 (rank : 15) | θ value | 0.0563607 (rank : 13) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7Z6W7, Q5H904, Q8WYJ7 | Gene names | DNAJB7, HSC3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 7. | |||||
|
DNJB8_MOUSE
|
||||||
| NC score | 0.144030 (rank : 16) | θ value | 0.0961366 (rank : 19) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QYI7 | Gene names | Dnajb8 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 8 (mDJ6). | |||||
|
DNJB8_HUMAN
|
||||||
| NC score | 0.136809 (rank : 17) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8NHS0 | Gene names | DNAJB8 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 8. | |||||
|
DNJB6_MOUSE
|
||||||
| NC score | 0.133283 (rank : 18) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O54946, Q99LA5, Q9QYI9 | Gene names | Dnajb6, Hsj2, Mrj | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MRJ) (mDj4). | |||||
|
DNJB6_HUMAN
|
||||||
| NC score | 0.133236 (rank : 19) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75190, O95806, Q9UIK6 | Gene names | DNAJB6, HSJ2, MSJ1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 6 (Heat shock protein J2) (HSJ-2) (MSJ-1) (HHDJ1) (MRJ). | |||||
|
DNJB1_HUMAN
|
||||||
| NC score | 0.128751 (rank : 20) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P25685 | Gene names | DNAJB1, DNAJ1, HDJ1, HSPF1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40) (DnaJ protein homolog 1) (HDJ-1). | |||||
|
DNJB5_MOUSE
|
||||||
| NC score | 0.126529 (rank : 21) | θ value | 0.125558 (rank : 20) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O89114 | Gene names | Dnajb5, Hsc40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40). | |||||
|
DNJBB_MOUSE
|
||||||
| NC score | 0.124713 (rank : 22) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99KV1, Q543I7 | Gene names | Dnajb11 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor. | |||||
|
DNJB2_HUMAN
|
||||||
| NC score | 0.123633 (rank : 23) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P25686, Q8IUK1, Q8IUK2, Q96F52 | Gene names | DNAJB2, HSJ1, HSPF3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 2 (Heat shock 40 kDa protein 3) (DnaJ protein homolog 1) (HSJ-1). | |||||
|
DNJB1_MOUSE
|
||||||
| NC score | 0.123391 (rank : 24) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QYJ3 | Gene names | Dnajb1, Hsp40, Hspf1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) (Heat shock protein 40) (HSP40). | |||||
|
DNJB5_HUMAN
|
||||||
| NC score | 0.123309 (rank : 25) | θ value | 0.163984 (rank : 22) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75953, Q8TDR7 | Gene names | DNAJB5, HSC40 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 5 (Heat shock protein Hsp40-3) (Heat shock protein cognate 40) (Hsc40) (Hsp40-2). | |||||
|
DNJBA_MOUSE
|
||||||
| NC score | 0.121317 (rank : 26) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QYI5 | Gene names | Dnajb10 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 10 (mDJ8). | |||||
|
DNJA4_HUMAN
|
||||||
| NC score | 0.121135 (rank : 27) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WW22, Q8N7P2 | Gene names | DNAJA4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4. | |||||
|
DNJB4_MOUSE
|
||||||
| NC score | 0.121070 (rank : 28) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9D832, Q3TS92, Q9D9U2 | Gene names | Dnajb4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4. | |||||
|
DNJA4_MOUSE
|
||||||
| NC score | 0.120850 (rank : 29) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JMC3, Q543S9, Q9CTD6, Q9CUD4 | Gene names | Dnaja4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 4 (MmDjA4). | |||||
|
DNJBB_HUMAN
|
||||||
| NC score | 0.119266 (rank : 30) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UBS4, Q96JC6 | Gene names | DNAJB11, EDJ, ERJ3 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 11 precursor (ER-associated dnaJ protein 3) (ErJ3) (ER-associated Hsp40 co-chaperone) (hDj9) (PWP1- interacting protein 4). | |||||
|
DJC17_MOUSE
|
||||||
| NC score | 0.113074 (rank : 31) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q91WT4, Q8C5R1 | Gene names | Dnajc17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DNJB4_HUMAN
|
||||||
| NC score | 0.112532 (rank : 32) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UDY4, Q13431 | Gene names | DNAJB4, DNAJW, HLJ1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 4 (Heat shock 40 kDa protein 1 homolog) (Heat shock protein 40 homolog) (HSP40 homolog). | |||||
|
DNJA1_HUMAN
|
||||||
| NC score | 0.111691 (rank : 33) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P31689 | Gene names | DNAJA1, DNAJ2, HDJ2, HSJ2, HSPF4 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2) (HSDJ). | |||||
|
DNJA1_MOUSE
|
||||||
| NC score | 0.111690 (rank : 34) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P63037, P54102 | Gene names | Dnaja1, Dnaj2, Hsj2, Hspf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily A member 1 (Heat shock 40 kDa protein 4) (DnaJ protein homolog 2) (HSJ-2). | |||||
|
DCJ11_HUMAN
|
||||||
| NC score | 0.110012 (rank : 35) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NVH1, Q4VWF5, Q5VZN0, Q6PK20, Q6PK70, Q8NDM2, Q96CL7 | Gene names | DNAJC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DCJ11_MOUSE
|
||||||
| NC score | 0.109986 (rank : 36) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5U458, Q8BP83, Q8C1Z4, Q8C6U5 | Gene names | Dnajc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 11. | |||||
|
DJC17_HUMAN
|
||||||
| NC score | 0.109277 (rank : 37) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NVM6 | Gene names | DNAJC17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DNJB3_MOUSE
|
||||||
| NC score | 0.108048 (rank : 38) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O35723, Q9DAN3, Q9DAN4 | Gene names | Dnajb3, Hsj3, Msj1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 3 (DnaJ protein homolog 3) (Heat shock J3 protein) (HSJ-3) (MSJ-1). | |||||
|
DNJ5G_HUMAN
|
||||||
| NC score | 0.106678 (rank : 39) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8N7S2, Q8IYQ4, Q96RJ8 | Gene names | DNAJC5G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 5G (Gamma cysteine string protein) (Gamma-CSP). | |||||
|
DNJC8_HUMAN
|
||||||
| NC score | 0.098737 (rank : 40) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75937, Q6IBA4, Q8N4Z5, Q9P051, Q9P067 | Gene names | DNAJC8, SPF31 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 8 (Splicing protein spf31). | |||||
|
DNJC8_MOUSE
|
||||||
| NC score | 0.098079 (rank : 41) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6NZB0, Q6PG72, Q8C2M6 | Gene names | Dnajc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 8. | |||||
|
DNJBD_HUMAN
|
||||||
| NC score | 0.096373 (rank : 42) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P59910, Q8IZW5 | Gene names | DNAJB13, TSARG3, TSARG6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DCJ14_HUMAN
|
||||||
| NC score | 0.096199 (rank : 43) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6Y2X3, Q66K17, Q96N59, Q96T63 | Gene names | DNAJC14, DRIP78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14 (Dopamine receptor-interacting protein of 78 kDa) (DRiP78). | |||||
|
DNJBD_MOUSE
|
||||||
| NC score | 0.095503 (rank : 44) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q80Y75, Q8CJA2 | Gene names | Dnajb13, Tsarg, Tsarg3, Tsarg6 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 13 (Testis spermatocyte apoptosis- related gene 6 protein) (Testis and spermatogenesis cell-related protein 6) (Testis spermatogenesis apoptosis-related gene 6 protein) (Testis spermatogenesis apoptosis-related gene 3 protein). | |||||
|
DNJC7_HUMAN
|
||||||
| NC score | 0.095094 (rank : 45) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99615 | Gene names | DNAJC7, TPR2, TTC2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2). | |||||
|
ZCSL3_HUMAN
|
||||||
| NC score | 0.094956 (rank : 46) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6P3W2 | Gene names | ZCSL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3. | |||||
|
DNJC7_MOUSE
|
||||||
| NC score | 0.094868 (rank : 47) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QYI3, Q3TKR1, Q8BPG3, Q8CIL2, Q9CT29, Q9D026 | Gene names | Dnajc7, Ttc2 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 7 (Tetratricopeptide repeat protein 2) (TPR repeat protein 2) (MDj11). | |||||
|
DNJA2_HUMAN
|
||||||
| NC score | 0.094744 (rank : 48) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O60884, O14711 | Gene names | DNAJA2, CPR3, HIRIP4 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily A member 2 (HIRA-interacting protein 4) (Cell cycle progression restoration gene 3 protein) (Dnj3) (NY-REN-14 antigen). | |||||
|
DNJA2_MOUSE
|
||||||
| NC score | 0.094676 (rank : 49) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QYJ0 | Gene names | Dnaja2 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily A member 2 (mDj3). | |||||
|
DNJB9_HUMAN
|
||||||
| NC score | 0.091799 (rank : 50) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UBS3 | Gene names | DNAJB9, MDG1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 9 (Microvascular endothelial differentiation gene 1 protein) (Mdg-1). | |||||
|
DNJB9_MOUSE
|
||||||
| NC score | 0.091689 (rank : 51) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QYI6, Q9DAW1 | Gene names | Dnajb9 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 9 (mDJ7). | |||||
|
DCJ14_MOUSE
|
||||||
| NC score | 0.091475 (rank : 52) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q921R4, Q3TX73, Q8BUU3, Q9CYB7 | Gene names | Dnajc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14. | |||||
|
DNJA3_MOUSE
|
||||||
| NC score | 0.089423 (rank : 53) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99M87, Q8BSM0, Q99L09, Q99P71, Q99P76, Q9CT11, Q9DBJ7, Q9DC44 | Gene names | Dnaja3, Tid1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (mTid-1). | |||||
|
DNJA3_HUMAN
|
||||||
| NC score | 0.088919 (rank : 54) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96EY1, O75472, Q8WUJ6, Q8WXJ3, Q96D76, Q96IV1, Q9NYH8 | Gene names | DNAJA3, TID1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (hTid-1). | |||||
|
DNJBC_MOUSE
|
||||||
| NC score | 0.088472 (rank : 55) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QYI4 | Gene names | Dnajb12 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 12 (mDJ10). | |||||
|
ZCSL3_MOUSE
|
||||||
| NC score | 0.087743 (rank : 56) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q91ZF0, Q9D9S7 | Gene names | Zcsl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CSL-type zinc finger-containing protein 3 (J-domain protein DjC7). | |||||
|
WBS18_MOUSE
|
||||||
| NC score | 0.087259 (rank : 57) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P59041 | Gene names | Wbscr18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein homolog. | |||||
|
DNJBC_HUMAN
|
||||||
| NC score | 0.086675 (rank : 58) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NXW2, Q9H6H0 | Gene names | DNAJB12 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily B member 12. | |||||
|
WBS18_HUMAN
|
||||||
| NC score | 0.085463 (rank : 59) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96LL9, Q9BSG8 | Gene names | WBSCR18 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 18 protein. | |||||
|
DJC18_HUMAN
|
||||||
| NC score | 0.085200 (rank : 60) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H819 | Gene names | DNAJC18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DNJC9_HUMAN
|
||||||
| NC score | 0.083384 (rank : 61) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WXX5 | Gene names | DNAJC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9 (DnaJ protein SB73). | |||||
|
DJC12_HUMAN
|
||||||
| NC score | 0.082631 (rank : 62) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UKB3, Q9UKB2 | Gene names | DNAJC12, JDP1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
DJC18_MOUSE
|
||||||
| NC score | 0.081553 (rank : 63) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9CZJ9, Q8R371 | Gene names | Dnajc18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 18. | |||||
|
DNJC4_HUMAN
|
||||||
| NC score | 0.081232 (rank : 64) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NNZ3, O14716 | Gene names | DNAJC4, HSPF2, MCG18 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18) (DnaJ-like protein HSPF2). | |||||
|
DNJC9_MOUSE
|
||||||
| NC score | 0.080810 (rank : 65) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91WN1, Q3TSG3, Q8R0E3 | Gene names | Dnajc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 9. | |||||
|
DJC16_HUMAN
|
||||||
| NC score | 0.080770 (rank : 66) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y2G8, Q68D57, Q86X32, Q8N5P4 | Gene names | DNAJC16, KIAA0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DNJC1_MOUSE
|
||||||
| NC score | 0.080349 (rank : 67) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61712 | Gene names | Dnajc1, Dnajl1, Mtj1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1 (DnaJ protein homolog MTJ1). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.080276 (rank : 68) | θ value | 0.0193708 (rank : 11) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
DNJC1_HUMAN
|
||||||
| NC score | 0.079759 (rank : 69) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96KC8 | Gene names | DNAJC1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1. | |||||
|
DJC16_MOUSE
|
||||||
| NC score | 0.079560 (rank : 70) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80TN4, Q811G1, Q8BHI2 | Gene names | Dnajc16, Kiaa0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
DNJC4_MOUSE
|
||||||
| NC score | 0.079125 (rank : 71) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D844, O70278 | Gene names | Dnajc4, Mcg18 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 4 (Multiple endocrine neoplasia type 1 candidate protein number 18 homolog). | |||||
|
AAK1_MOUSE
|
||||||
| NC score | 0.075006 (rank : 72) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
DJC12_MOUSE
|
||||||
| NC score | 0.074862 (rank : 73) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9R022 | Gene names | Dnajc12, Jdp1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
AAK1_HUMAN
|
||||||
| NC score | 0.074820 (rank : 74) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1077 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q2M2I8, Q4ZFZ3, Q53RX6, Q9UPV4 | Gene names | AAK1, KIAA1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
BMP2K_MOUSE
|
||||||
| NC score | 0.073136 (rank : 75) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91Z96, Q8C8L7 | Gene names | Bmp2k, Bike | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
ZRF1_HUMAN
|
||||||
| NC score | 0.072552 (rank : 76) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.072266 (rank : 77) | θ value | 0.125558 (rank : 21) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
DNJC3_HUMAN
|
||||||
| NC score | 0.072253 (rank : 78) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13217, Q86WT9, Q8N4N2 | Gene names | DNAJC3, P58IPK, PRKRI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
BMP2K_HUMAN
|
||||||
| NC score | 0.072224 (rank : 79) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NSY1, O94791, Q8IYF2, Q8N2G7, Q8NHG9, Q9NTG8 | Gene names | BMP2K, BIKE | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
DNJC3_MOUSE
|
||||||
| NC score | 0.071930 (rank : 80) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91YW3, Q60873 | Gene names | Dnajc3, P58ipk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 3 (Interferon-induced, double-stranded RNA-activated protein kinase inhibitor) (Protein kinase inhibitor p58) (Protein kinase inhibitor of 58 kDa). | |||||
|
ZRF1_MOUSE
|
||||||
| NC score | 0.071284 (rank : 81) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
TENS4_MOUSE
|
||||||
| NC score | 0.069771 (rank : 82) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BZ33, Q3TCM8, Q7TNR5 | Gene names | Tns4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor. | |||||
|
TENS4_HUMAN
|
||||||
| NC score | 0.068548 (rank : 83) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IZW8, Q71RB7, Q8WV64, Q96JV4 | Gene names | TNS4, CTEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor (C-terminal tensin-like protein). | |||||
|
PDXL2_MOUSE
|
||||||
| NC score | 0.061157 (rank : 84) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.058314 (rank : 85) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.058059 (rank : 86) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
K2002_HUMAN
|
||||||
| NC score | 0.057238 (rank : 87) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H792, Q6ZS78, Q8NCM3 | Gene names | KIAA2002 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized tyrosine-protein kinase KIAA2002 (EC 2.7.10.2). | |||||
|
DNJCD_HUMAN
|
||||||
| NC score | 0.053262 (rank : 88) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75165, Q6PI82, Q6UJ77, Q6ZSW1, Q6ZUT5, Q86XG3, Q96DC1, Q9BWK9 | Gene names | DNAJC13, KIAA0678, RME8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 13 (Required for receptor-mediated endocytosis 8). | |||||
|
CU055_MOUSE
|
||||||
| NC score | 0.052172 (rank : 89) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VCE1, Q99JX0 | Gene names | ORF28 | |||
|
Domain Architecture |

|
|||||
| Description | J domain-containing protein C21orf55 homolog. | |||||
|
PRP5_HUMAN
|
||||||
| NC score | 0.050796 (rank : 90) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
DCBD1_MOUSE
|
||||||
| NC score | 0.050273 (rank : 91) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D4J3, Q8R327, Q9D696 | Gene names | Dcbld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.046993 (rank : 92) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
TNR6A_HUMAN
|
||||||
| NC score | 0.043987 (rank : 93) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
TTLL5_MOUSE
|
||||||
| NC score | 0.039842 (rank : 94) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
HRBL_HUMAN
|
||||||
| NC score | 0.036369 (rank : 95) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.034901 (rank : 96) | θ value | 0.0563607 (rank : 14) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
INT1_MOUSE
|
||||||
| NC score | 0.034152 (rank : 97) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P4S8, Q80UQ7, Q91Z01, Q9CTF7 | Gene names | Ints1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 1 (Int1). | |||||
|
CTND2_HUMAN
|
||||||
| NC score | 0.032042 (rank : 98) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.030814 (rank : 99) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.028007 (rank : 100) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PAX9_HUMAN
|
||||||
| NC score | 0.027646 (rank : 101) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55771, Q99582, Q9UQR4 | Gene names | PAX9 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-9. | |||||
|
PAX9_MOUSE
|
||||||
| NC score | 0.027376 (rank : 102) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47242 | Gene names | Pax9, Pax-9 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-9. | |||||
|
CTND2_MOUSE
|
||||||
| NC score | 0.026801 (rank : 103) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
GATA5_MOUSE
|
||||||
| NC score | 0.025884 (rank : 104) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97489 | Gene names | Gata5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
COE3_HUMAN
|
||||||
| NC score | 0.023251 (rank : 105) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H4W6, Q5T6H9, Q9H4W5 | Gene names | EBF3, COE3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
TTLL5_HUMAN
|
||||||
| NC score | 0.023101 (rank : 106) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6EMB2, Q9P1V5, Q9UPZ4 | Gene names | TTLL5, KIAA0998, STAMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5 (SRC1 and TIF2-associated modulatory protein). | |||||
|
JIP1_HUMAN
|
||||||
| NC score | 0.022923 (rank : 107) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
HRBL_MOUSE
|
||||||
| NC score | 0.022039 (rank : 108) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
COE3_MOUSE
|
||||||
| NC score | 0.021870 (rank : 109) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08791, O08793 | Gene names | Ebf3, Coe3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
MGRN1_HUMAN
|
||||||
| NC score | 0.021651 (rank : 110) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60291, Q86W76 | Gene names | MGRN1, KIAA0544, RNF156 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase MGRN1 (EC 6.3.2.-) (Mahogunin ring finger protein 1) (RING finger protein 156). | |||||
|
INT1_HUMAN
|
||||||
| NC score | 0.021202 (rank : 111) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N201, Q6NT70, Q6UX74, Q8WV40, Q96D36, Q9NTD1, Q9P2A8, Q9Y3W8 | Gene names | INTS1, KIAA1440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 1 (Int1). | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.020272 (rank : 112) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
T22D4_HUMAN
|
||||||
| NC score | 0.019067 (rank : 113) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
LPIN3_HUMAN
|
||||||
| NC score | 0.017110 (rank : 114) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQK8, Q5TDB9, Q9NPY8, Q9UJE5 | Gene names | LPIN3, LIPN3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-3 (Lipin 3-like). | |||||
|
TRIP6_HUMAN
|
||||||
| NC score | 0.016805 (rank : 115) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15654, O15170, O15275, Q9BTB2, Q9UNT4 | Gene names | TRIP6, OIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 6 (TRIP6) (OPA-interacting protein 1) (Zyxin-related protein 1) (ZRP-1). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.016668 (rank : 116) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.016625 (rank : 117) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
CT151_HUMAN
|
||||||
| NC score | 0.015717 (rank : 118) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NC74, Q8N4Z9, Q9BR75, Q9H0Y9 | Gene names | C20orf151 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf151. | |||||
|
FOXI1_HUMAN
|
||||||
| NC score | 0.014600 (rank : 119) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12951, Q14518, Q66SR7, Q8N6L8 | Gene names | FOXI1, FKHL10, FREAC6 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein I1 (Forkhead-related protein FKHL10) (Forkhead- related transcription factor 6) (FREAC-6) (Hepatocyte nuclear factor 3 forkhead homolog 3) (HNF-3/fork-head homolog 3) (HFH-3). | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | 0.014490 (rank : 120) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.014353 (rank : 121) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.014066 (rank : 122) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
TACC3_HUMAN
|
||||||
| NC score | 0.014036 (rank : 123) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
AATK_HUMAN
|
||||||
| NC score | 0.013732 (rank : 124) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
DAXX_MOUSE
|
||||||
| NC score | 0.013643 (rank : 125) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35613, Q9QWT8, Q9QWV3 | Gene names | Daxx | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx). | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.013251 (rank : 126) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
ATS14_HUMAN
|
||||||
| NC score | 0.013112 (rank : 127) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WXS8, Q8TE55, Q8TEY8 | Gene names | ADAMTS14 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-14 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 14) (ADAM-TS 14) (ADAM-TS14). | |||||
|
JIP1_MOUSE
|
||||||
| NC score | 0.012931 (rank : 128) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
CAD11_HUMAN
|
||||||
| NC score | 0.011940 (rank : 129) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55287, Q15065, Q15066, Q9UQ93, Q9UQ94 | Gene names | CDH11 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-11 precursor (Osteoblast-cadherin) (OB-cadherin) (OSF-4). | |||||
|
CAD11_MOUSE
|
||||||
| NC score | 0.011937 (rank : 130) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55288 | Gene names | Cdh11, Cad-11 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-11 precursor (Osteoblast-cadherin) (OB-cadherin) (OSF-4). | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.011075 (rank : 131) | θ value | 0.0736092 (rank : 18) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.009781 (rank : 132) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.008498 (rank : 133) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
BNC1_HUMAN
|
||||||
| NC score | 0.006961 (rank : 134) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01954, Q15840 | Gene names | BNC1, BNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-1. | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.006728 (rank : 135) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CAD15_HUMAN
|
||||||
| NC score | 0.006662 (rank : 136) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55291 | Gene names | CDH15, CDH14, CDH3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-15 precursor (Muscle-cadherin) (M-cadherin) (Cadherin-14). | |||||
|
STAT6_MOUSE
|
||||||
| NC score | 0.005767 (rank : 137) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52633 | Gene names | Stat6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and transcription activator 6. | |||||
|
GRIP2_HUMAN
|
||||||
| NC score | 0.005035 (rank : 138) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||