Please be patient as the page loads
|
P3H1_MOUSE
|
||||||
| SwissProt Accessions | Q3V1T4, Q3TWX8, Q8BSV2, Q8CFL3, Q9CWK5, Q9QZT6, Q9QZT7 | Gene names | Lepre1, Gros1, P3h1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 1 precursor (EC 1.14.11.7) (Leucine- and proline- enriched proteoglycan 1) (Leprecan-1) (Growth suppressor 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
P3H1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998206 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q32P28, Q7KZR4, Q96BR8, Q96SK8, Q96SL5, Q96SN3, Q9H6K3, Q9HC86, Q9HC87 | Gene names | LEPRE1, GROS1, P3H1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 1 precursor (EC 1.14.11.7) (Leucine- and proline- enriched proteoglycan 1) (Leprecan-1) (Growth suppressor 1). | |||||
|
P3H1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q3V1T4, Q3TWX8, Q8BSV2, Q8CFL3, Q9CWK5, Q9QZT6, Q9QZT7 | Gene names | Lepre1, Gros1, P3h1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 1 precursor (EC 1.14.11.7) (Leucine- and proline- enriched proteoglycan 1) (Leprecan-1) (Growth suppressor 1). | |||||
|
P3H2_HUMAN
|
||||||
| θ value | 1.60882e-182 (rank : 3) | NC score | 0.983680 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IVL5, Q9NVI2 | Gene names | LEPREL1, MLAT4, P3H2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 2 precursor (EC 1.14.11.7) (Leprecan-like protein 1) (Myxoid liposarcoma-associated protein 4). | |||||
|
P3H2_MOUSE
|
||||||
| θ value | 2.32313e-181 (rank : 4) | NC score | 0.983182 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CG71, Q8C673 | Gene names | Leprel1, P3h2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 2 precursor (EC 1.14.11.7) (Leprecan-like protein 1). | |||||
|
P3H3_HUMAN
|
||||||
| θ value | 1.47209e-143 (rank : 5) | NC score | 0.969607 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IVL6, Q13512, Q15740, Q66K32, Q6NX61, Q7L2T1 | Gene names | LEPREL2, P3H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B). | |||||
|
P3H3_MOUSE
|
||||||
| θ value | 1.20704e-137 (rank : 6) | NC score | 0.967732 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CG70, O88836 | Gene names | Leprel2, P3h3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B). | |||||
|
CRTAP_HUMAN
|
||||||
| θ value | 2.94719e-51 (rank : 7) | NC score | 0.876571 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75718 | Gene names | CRTAP, CASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage-associated protein precursor. | |||||
|
CRTAP_MOUSE
|
||||||
| θ value | 1.67352e-46 (rank : 8) | NC score | 0.867512 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CYD3, O88698 | Gene names | Crtap, Casp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage-associated protein precursor. | |||||
|
NO55_HUMAN
|
||||||
| θ value | 1.19932e-44 (rank : 9) | NC score | 0.860612 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92791, Q9H4F6 | Gene names | SC65, NOL55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar autoantigen No55. | |||||
|
P4HA2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 10) | NC score | 0.116125 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60716, Q8VBU4 | Gene names | P4ha2 | |||
|
Domain Architecture |

|
|||||
| Description | Prolyl 4-hydroxylase alpha-2 subunit precursor (EC 1.14.11.2) (4-PH alpha-2) (Procollagen-proline,2-oxoglutarate-4-dioxygenase alpha-2 subunit). | |||||
|
EGLN2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.090256 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91YE2, Q8C6I4, Q8CIL9, Q8VHJ1, Q99MI0 | Gene names | Egln2 | |||
|
Domain Architecture |
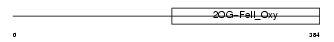
|
|||||
| Description | Egl nine homolog 2 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 1) (HIF-prolyl hydroxylase 1) (HIF-PH1) (Falkor). | |||||
|
P4HA2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.091725 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15460, Q8WWN0 | Gene names | P4HA2 | |||
|
Domain Architecture |

|
|||||
| Description | Prolyl 4-hydroxylase alpha-2 subunit precursor (EC 1.14.11.2) (4-PH alpha-2) (Procollagen-proline,2-oxoglutarate-4-dioxygenase alpha-2 subunit). | |||||
|
NOP14_HUMAN
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.046570 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
EGLN1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 14) | NC score | 0.082320 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9GZT9, Q8N3M8, Q9BZS8, Q9BZT0 | Gene names | EGLN1, C1orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (Prolyl hydroxylase domain-containing protein 2) (PHD2) (SM-20). | |||||
|
EGLN1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.081157 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91YE3, Q8VHJ2, Q922P3 | Gene names | Egln1 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (SM-20). | |||||
|
EGLN2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.080928 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96KS0, Q8WWY4, Q9BV14 | Gene names | EGLN2, EIT6 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 2 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 1) (HIF-prolyl hydroxylase 1) (HIF-PH1) (HPH-3) (Prolyl hydroxylase domain-containing protein 1) (PHD1) (Estrogen-induced tag 6). | |||||
|
NOP14_MOUSE
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.045672 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R3N1 | Gene names | Nop14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.019077 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
ACAD8_MOUSE
|
||||||
| θ value | 1.38821 (rank : 19) | NC score | 0.012410 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D7B6, Q8BK36, Q8BKQ8, Q8CFY9, Q9D2L8 | Gene names | Acad8 | |||
|
Domain Architecture |
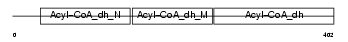
|
|||||
| Description | Acyl-CoA dehydrogenase family member 8, mitochondrial precursor (EC 1.3.99.-) (ACAD-8) (Isobutyryl-CoA dehydrogenase). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 2.36792 (rank : 20) | NC score | 0.012822 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
P4HA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.075340 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P13674, Q15082, Q15083, Q5VSQ5 | Gene names | P4HA1, P4HA | |||
|
Domain Architecture |

|
|||||
| Description | Prolyl 4-hydroxylase alpha-1 subunit precursor (EC 1.14.11.2) (4-PH alpha-1) (Procollagen-proline,2-oxoglutarate-4-dioxygenase alpha-1 subunit). | |||||
|
UN45A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.013171 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
UACA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.003506 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
UBR2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.010338 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6WKZ8, Q6DIB9, Q80U31, Q8BUL9, Q8CGW0, Q8K2I6, Q8R0V7, Q8R130 | Gene names | Ubr2, Kiaa0349 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-2 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-2) (Ubiquitin-protein ligase E3- alpha-II). | |||||
|
EGLN3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.060537 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H6Z9 | Gene names | EGLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 3 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 3) (HIF-prolyl hydroxylase 3) (HIF-PH3) (HPH-1) (Prolyl hydroxylase domain-containing protein 3) (PHD3). | |||||
|
EGLN3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.060555 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91UZ4, Q3TCG8, Q8C8M6, Q8CCA8, Q8R5C7 | Gene names | Egln3 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 3 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 3) (HIF-prolyl hydroxylase 3) (HIF-PH3) (SM-20). | |||||
|
NP1L3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.009368 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
BUB1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.009423 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1S0 | Gene names | Bub1b, Mad3l | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.001019 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.001901 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.003307 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
UN45A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.011669 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
4ET_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.014348 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NRA8, Q8NCF2, Q9H708 | Gene names | EIF4ENIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
CENPJ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.005903 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | -0.001122 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | -0.001226 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
P4HA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.070432 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60715, Q3TEB7, Q80T05, Q91VJ7 | Gene names | P4ha1 | |||
|
Domain Architecture |

|
|||||
| Description | Prolyl 4-hydroxylase alpha-1 subunit precursor (EC 1.14.11.2) (4-PH alpha-1) (Procollagen-proline,2-oxoglutarate-4-dioxygenase alpha-1 subunit). | |||||
|
P3H1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q3V1T4, Q3TWX8, Q8BSV2, Q8CFL3, Q9CWK5, Q9QZT6, Q9QZT7 | Gene names | Lepre1, Gros1, P3h1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 1 precursor (EC 1.14.11.7) (Leucine- and proline- enriched proteoglycan 1) (Leprecan-1) (Growth suppressor 1). | |||||
|
P3H1_HUMAN
|
||||||
| NC score | 0.998206 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q32P28, Q7KZR4, Q96BR8, Q96SK8, Q96SL5, Q96SN3, Q9H6K3, Q9HC86, Q9HC87 | Gene names | LEPRE1, GROS1, P3H1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 1 precursor (EC 1.14.11.7) (Leucine- and proline- enriched proteoglycan 1) (Leprecan-1) (Growth suppressor 1). | |||||
|
P3H2_HUMAN
|
||||||
| NC score | 0.983680 (rank : 3) | θ value | 1.60882e-182 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IVL5, Q9NVI2 | Gene names | LEPREL1, MLAT4, P3H2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 2 precursor (EC 1.14.11.7) (Leprecan-like protein 1) (Myxoid liposarcoma-associated protein 4). | |||||
|
P3H2_MOUSE
|
||||||
| NC score | 0.983182 (rank : 4) | θ value | 2.32313e-181 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CG71, Q8C673 | Gene names | Leprel1, P3h2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 2 precursor (EC 1.14.11.7) (Leprecan-like protein 1). | |||||
|
P3H3_HUMAN
|
||||||
| NC score | 0.969607 (rank : 5) | θ value | 1.47209e-143 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IVL6, Q13512, Q15740, Q66K32, Q6NX61, Q7L2T1 | Gene names | LEPREL2, P3H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B). | |||||
|
P3H3_MOUSE
|
||||||
| NC score | 0.967732 (rank : 6) | θ value | 1.20704e-137 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CG70, O88836 | Gene names | Leprel2, P3h3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B). | |||||
|
CRTAP_HUMAN
|
||||||
| NC score | 0.876571 (rank : 7) | θ value | 2.94719e-51 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75718 | Gene names | CRTAP, CASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage-associated protein precursor. | |||||
|
CRTAP_MOUSE
|
||||||
| NC score | 0.867512 (rank : 8) | θ value | 1.67352e-46 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CYD3, O88698 | Gene names | Crtap, Casp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage-associated protein precursor. | |||||
|
NO55_HUMAN
|
||||||
| NC score | 0.860612 (rank : 9) | θ value | 1.19932e-44 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92791, Q9H4F6 | Gene names | SC65, NOL55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar autoantigen No55. | |||||
|
P4HA2_MOUSE
|
||||||
| NC score | 0.116125 (rank : 10) | θ value | 0.0113563 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60716, Q8VBU4 | Gene names | P4ha2 | |||
|
Domain Architecture |

|
|||||
| Description | Prolyl 4-hydroxylase alpha-2 subunit precursor (EC 1.14.11.2) (4-PH alpha-2) (Procollagen-proline,2-oxoglutarate-4-dioxygenase alpha-2 subunit). | |||||
|
P4HA2_HUMAN
|
||||||
| NC score | 0.091725 (rank : 11) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15460, Q8WWN0 | Gene names | P4HA2 | |||
|
Domain Architecture |

|
|||||
| Description | Prolyl 4-hydroxylase alpha-2 subunit precursor (EC 1.14.11.2) (4-PH alpha-2) (Procollagen-proline,2-oxoglutarate-4-dioxygenase alpha-2 subunit). | |||||
|
EGLN2_MOUSE
|
||||||
| NC score | 0.090256 (rank : 12) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91YE2, Q8C6I4, Q8CIL9, Q8VHJ1, Q99MI0 | Gene names | Egln2 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 2 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 1) (HIF-prolyl hydroxylase 1) (HIF-PH1) (Falkor). | |||||
|
EGLN1_HUMAN
|
||||||
| NC score | 0.082320 (rank : 13) | θ value | 0.813845 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9GZT9, Q8N3M8, Q9BZS8, Q9BZT0 | Gene names | EGLN1, C1orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (Prolyl hydroxylase domain-containing protein 2) (PHD2) (SM-20). | |||||
|
EGLN1_MOUSE
|
||||||
| NC score | 0.081157 (rank : 14) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91YE3, Q8VHJ2, Q922P3 | Gene names | Egln1 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (SM-20). | |||||
|
EGLN2_HUMAN
|
||||||
| NC score | 0.080928 (rank : 15) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96KS0, Q8WWY4, Q9BV14 | Gene names | EGLN2, EIT6 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 2 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 1) (HIF-prolyl hydroxylase 1) (HIF-PH1) (HPH-3) (Prolyl hydroxylase domain-containing protein 1) (PHD1) (Estrogen-induced tag 6). | |||||
|
P4HA1_HUMAN
|
||||||
| NC score | 0.075340 (rank : 16) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P13674, Q15082, Q15083, Q5VSQ5 | Gene names | P4HA1, P4HA | |||
|
Domain Architecture |

|
|||||
| Description | Prolyl 4-hydroxylase alpha-1 subunit precursor (EC 1.14.11.2) (4-PH alpha-1) (Procollagen-proline,2-oxoglutarate-4-dioxygenase alpha-1 subunit). | |||||
|
P4HA1_MOUSE
|
||||||
| NC score | 0.070432 (rank : 17) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60715, Q3TEB7, Q80T05, Q91VJ7 | Gene names | P4ha1 | |||
|
Domain Architecture |

|
|||||
| Description | Prolyl 4-hydroxylase alpha-1 subunit precursor (EC 1.14.11.2) (4-PH alpha-1) (Procollagen-proline,2-oxoglutarate-4-dioxygenase alpha-1 subunit). | |||||
|
EGLN3_MOUSE
|
||||||
| NC score | 0.060555 (rank : 18) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91UZ4, Q3TCG8, Q8C8M6, Q8CCA8, Q8R5C7 | Gene names | Egln3 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 3 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 3) (HIF-prolyl hydroxylase 3) (HIF-PH3) (SM-20). | |||||
|
EGLN3_HUMAN
|
||||||
| NC score | 0.060537 (rank : 19) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H6Z9 | Gene names | EGLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 3 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 3) (HIF-prolyl hydroxylase 3) (HIF-PH3) (HPH-1) (Prolyl hydroxylase domain-containing protein 3) (PHD3). | |||||
|
NOP14_HUMAN
|
||||||
| NC score | 0.046570 (rank : 20) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
NOP14_MOUSE
|
||||||
| NC score | 0.045672 (rank : 21) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R3N1 | Gene names | Nop14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.019077 (rank : 22) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
4ET_HUMAN
|
||||||
| NC score | 0.014348 (rank : 23) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NRA8, Q8NCF2, Q9H708 | Gene names | EIF4ENIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
UN45A_MOUSE
|
||||||
| NC score | 0.013171 (rank : 24) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.012822 (rank : 25) | θ value | 2.36792 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
ACAD8_MOUSE
|
||||||
| NC score | 0.012410 (rank : 26) | θ value | 1.38821 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D7B6, Q8BK36, Q8BKQ8, Q8CFY9, Q9D2L8 | Gene names | Acad8 | |||
|
Domain Architecture |
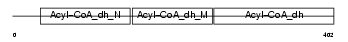
|
|||||
| Description | Acyl-CoA dehydrogenase family member 8, mitochondrial precursor (EC 1.3.99.-) (ACAD-8) (Isobutyryl-CoA dehydrogenase). | |||||
|
UN45A_HUMAN
|
||||||
| NC score | 0.011669 (rank : 27) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
UBR2_MOUSE
|
||||||
| NC score | 0.010338 (rank : 28) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6WKZ8, Q6DIB9, Q80U31, Q8BUL9, Q8CGW0, Q8K2I6, Q8R0V7, Q8R130 | Gene names | Ubr2, Kiaa0349 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3 component N-recognin-2 (EC 6.-.-.-) (Ubiquitin-protein ligase E3-alpha-2) (Ubiquitin-protein ligase E3- alpha-II). | |||||
|
BUB1B_MOUSE
|
||||||
| NC score | 0.009423 (rank : 29) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1S0 | Gene names | Bub1b, Mad3l | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L). | |||||
|
NP1L3_MOUSE
|
||||||
| NC score | 0.009368 (rank : 30) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
CENPJ_HUMAN
|
||||||
| NC score | 0.005903 (rank : 31) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
UACA_MOUSE
|
||||||
| NC score | 0.003506 (rank : 32) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.003307 (rank : 33) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.001901 (rank : 34) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.001019 (rank : 35) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | -0.001122 (rank : 36) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | -0.001226 (rank : 37) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||