Please be patient as the page loads
|
DDX24_HUMAN
|
||||||
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DDX24_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 190 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX24_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.992652 (rank : 2) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX47_HUMAN
|
||||||
| θ value | 3.72821e-46 (rank : 3) | NC score | 0.949644 (rank : 4) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX47_MOUSE
|
||||||
| θ value | 8.30557e-46 (rank : 4) | NC score | 0.949864 (rank : 3) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX49_HUMAN
|
||||||
| θ value | 1.79215e-40 (rank : 5) | NC score | 0.943132 (rank : 6) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX49_MOUSE
|
||||||
| θ value | 1.6774e-38 (rank : 6) | NC score | 0.945605 (rank : 5) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX18_HUMAN
|
||||||
| θ value | 1.46948e-34 (rank : 7) | NC score | 0.933609 (rank : 8) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
DDX18_MOUSE
|
||||||
| θ value | 2.50655e-34 (rank : 8) | NC score | 0.933474 (rank : 9) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
DDX27_MOUSE
|
||||||
| θ value | 9.52487e-34 (rank : 9) | NC score | 0.934666 (rank : 7) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX41_HUMAN
|
||||||
| θ value | 2.77131e-33 (rank : 10) | NC score | 0.932960 (rank : 11) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
DDX41_MOUSE
|
||||||
| θ value | 2.77131e-33 (rank : 11) | NC score | 0.932641 (rank : 13) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
DDX17_HUMAN
|
||||||
| θ value | 6.17384e-33 (rank : 12) | NC score | 0.926165 (rank : 21) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
DDX17_MOUSE
|
||||||
| θ value | 6.17384e-33 (rank : 13) | NC score | 0.926298 (rank : 20) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
DDX27_HUMAN
|
||||||
| θ value | 6.17384e-33 (rank : 14) | NC score | 0.932956 (rank : 12) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX54_MOUSE
|
||||||
| θ value | 1.52067e-31 (rank : 15) | NC score | 0.931930 (rank : 14) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
DDX10_HUMAN
|
||||||
| θ value | 5.77852e-31 (rank : 16) | NC score | 0.925860 (rank : 22) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX10_MOUSE
|
||||||
| θ value | 4.89182e-30 (rank : 17) | NC score | 0.925287 (rank : 23) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX54_HUMAN
|
||||||
| θ value | 8.3442e-30 (rank : 18) | NC score | 0.927540 (rank : 18) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
DDX55_HUMAN
|
||||||
| θ value | 1.08979e-29 (rank : 19) | NC score | 0.931458 (rank : 15) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX55_MOUSE
|
||||||
| θ value | 1.4233e-29 (rank : 20) | NC score | 0.933048 (rank : 10) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX5_HUMAN
|
||||||
| θ value | 5.40856e-29 (rank : 21) | NC score | 0.919632 (rank : 29) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
DDX5_MOUSE
|
||||||
| θ value | 5.40856e-29 (rank : 22) | NC score | 0.919611 (rank : 30) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
DDX46_HUMAN
|
||||||
| θ value | 5.97985e-28 (rank : 23) | NC score | 0.907071 (rank : 54) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX46_MOUSE
|
||||||
| θ value | 5.97985e-28 (rank : 24) | NC score | 0.905425 (rank : 55) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
DDX48_HUMAN
|
||||||
| θ value | 5.97985e-28 (rank : 25) | NC score | 0.918634 (rank : 31) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX48_MOUSE
|
||||||
| θ value | 5.97985e-28 (rank : 26) | NC score | 0.918482 (rank : 32) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
DDX43_HUMAN
|
||||||
| θ value | 3.87602e-27 (rank : 27) | NC score | 0.924561 (rank : 24) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
DDX23_HUMAN
|
||||||
| θ value | 6.61148e-27 (rank : 28) | NC score | 0.900346 (rank : 56) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
DDX56_HUMAN
|
||||||
| θ value | 6.61148e-27 (rank : 29) | NC score | 0.922389 (rank : 27) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NY93, Q9H9I8 | Gene names | DDX56, DDX21, NOH61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase) (DEAD-box protein 21). | |||||
|
DDX3Y_MOUSE
|
||||||
| θ value | 1.12775e-26 (rank : 30) | NC score | 0.911440 (rank : 47) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
DDX56_MOUSE
|
||||||
| θ value | 1.12775e-26 (rank : 31) | NC score | 0.921299 (rank : 28) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
DDX31_HUMAN
|
||||||
| θ value | 1.92365e-26 (rank : 32) | NC score | 0.930129 (rank : 16) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
DDX3X_HUMAN
|
||||||
| θ value | 1.92365e-26 (rank : 33) | NC score | 0.910277 (rank : 51) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX3X_MOUSE
|
||||||
| θ value | 4.28545e-26 (rank : 34) | NC score | 0.909567 (rank : 52) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
PL10_MOUSE
|
||||||
| θ value | 4.28545e-26 (rank : 35) | NC score | 0.910320 (rank : 50) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
DDX3Y_HUMAN
|
||||||
| θ value | 9.54697e-26 (rank : 36) | NC score | 0.907734 (rank : 53) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
DDX28_HUMAN
|
||||||
| θ value | 1.24688e-25 (rank : 37) | NC score | 0.926474 (rank : 19) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX28_MOUSE
|
||||||
| θ value | 1.24688e-25 (rank : 38) | NC score | 0.928753 (rank : 17) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
IF4A1_HUMAN
|
||||||
| θ value | 4.73814e-25 (rank : 39) | NC score | 0.912849 (rank : 42) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| θ value | 4.73814e-25 (rank : 40) | NC score | 0.912849 (rank : 43) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A2_HUMAN
|
||||||
| θ value | 4.73814e-25 (rank : 41) | NC score | 0.912483 (rank : 45) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| θ value | 4.73814e-25 (rank : 42) | NC score | 0.912483 (rank : 46) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
DDX39_HUMAN
|
||||||
| θ value | 1.05554e-24 (rank : 43) | NC score | 0.915274 (rank : 36) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O00148, Q9BVP6, Q9H5W0 | Gene names | DDX39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39) (Nuclear RNA helicase URH49). | |||||
|
DDX39_MOUSE
|
||||||
| θ value | 1.80048e-24 (rank : 44) | NC score | 0.914840 (rank : 37) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8VDW0, Q3UJV4, Q8C2C2 | Gene names | Ddx39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39). | |||||
|
DDX50_MOUSE
|
||||||
| θ value | 4.01107e-24 (rank : 45) | NC score | 0.911056 (rank : 48) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX4_HUMAN
|
||||||
| θ value | 5.23862e-24 (rank : 46) | NC score | 0.916946 (rank : 34) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
DDX50_HUMAN
|
||||||
| θ value | 5.23862e-24 (rank : 47) | NC score | 0.910536 (rank : 49) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX6_HUMAN
|
||||||
| θ value | 5.23862e-24 (rank : 48) | NC score | 0.923597 (rank : 25) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
DDX6_MOUSE
|
||||||
| θ value | 5.23862e-24 (rank : 49) | NC score | 0.923585 (rank : 26) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
DDX4_MOUSE
|
||||||
| θ value | 8.93572e-24 (rank : 50) | NC score | 0.917514 (rank : 33) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
UAP56_HUMAN
|
||||||
| θ value | 8.93572e-24 (rank : 51) | NC score | 0.913802 (rank : 39) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q13838, O43496, Q5RJ64, Q5RJ66, Q5ST94, Q5STB4, Q5STB5, Q5STB7, Q5STU4, Q5STU5, Q5STU6, Q5STU8, Q71V76 | Gene names | BAT1, UAP56 | |||
|
Domain Architecture |

|
|||||
| Description | Spliceosome RNA helicase BAT1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (ATP-dependent RNA helicase p47) (HLA-B-associated transcript-1). | |||||
|
UAP56_MOUSE
|
||||||
| θ value | 8.93572e-24 (rank : 52) | NC score | 0.913732 (rank : 40) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9Z1N5, Q8HW97 | Gene names | Bat1, Bat1a, Uap56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spliceosome RNA helicase Bat1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (HLA-B-associated transcript 1). | |||||
|
DDX53_HUMAN
|
||||||
| θ value | 1.5242e-23 (rank : 53) | NC score | 0.916856 (rank : 35) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
DDX51_MOUSE
|
||||||
| θ value | 4.43474e-23 (rank : 54) | NC score | 0.914404 (rank : 38) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 5.79196e-23 (rank : 55) | NC score | 0.899497 (rank : 57) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX1_MOUSE
|
||||||
| θ value | 3.75424e-22 (rank : 56) | NC score | 0.870999 (rank : 67) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 8.36355e-22 (rank : 57) | NC score | 0.892264 (rank : 61) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX1_HUMAN
|
||||||
| θ value | 1.09232e-21 (rank : 58) | NC score | 0.869761 (rank : 68) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
DDX51_HUMAN
|
||||||
| θ value | 1.86321e-21 (rank : 59) | NC score | 0.897591 (rank : 59) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX20_MOUSE
|
||||||
| θ value | 3.88503e-19 (rank : 60) | NC score | 0.896185 (rank : 60) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DDX20_HUMAN
|
||||||
| θ value | 6.62687e-19 (rank : 61) | NC score | 0.898933 (rank : 58) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
DDX52_HUMAN
|
||||||
| θ value | 2.13179e-17 (rank : 62) | NC score | 0.913060 (rank : 41) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX52_MOUSE
|
||||||
| θ value | 2.13179e-17 (rank : 63) | NC score | 0.912606 (rank : 44) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8K301, Q8BV29 | Gene names | Ddx52, Rok1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DD19A_HUMAN
|
||||||
| θ value | 2.35696e-16 (rank : 64) | NC score | 0.881676 (rank : 62) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19B_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 65) | NC score | 0.880638 (rank : 63) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
DD19A_MOUSE
|
||||||
| θ value | 1.52774e-15 (rank : 66) | NC score | 0.879955 (rank : 64) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DDX25_MOUSE
|
||||||
| θ value | 3.18553e-13 (rank : 67) | NC score | 0.877940 (rank : 66) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX25_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 68) | NC score | 0.878419 (rank : 65) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
WRN_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 69) | NC score | 0.468722 (rank : 73) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
RECQ1_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 70) | NC score | 0.434389 (rank : 74) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
DICER_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 71) | NC score | 0.402072 (rank : 80) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
DICER_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 72) | NC score | 0.401518 (rank : 81) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
WRN_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 73) | NC score | 0.429309 (rank : 77) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
BLM_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 74) | NC score | 0.473534 (rank : 72) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
BLM_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 75) | NC score | 0.484691 (rank : 71) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O88700, O88198 | Gene names | Blm | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein homolog (EC 3.6.1.-) (mBLM). | |||||
|
RECQ1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 76) | NC score | 0.429340 (rank : 76) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P46063 | Gene names | RECQL, RECQL1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
RECQ4_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 77) | NC score | 0.510439 (rank : 70) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
RECQ4_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 78) | NC score | 0.526881 (rank : 69) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
TTC16_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 79) | NC score | 0.034128 (rank : 96) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
FANCM_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 80) | NC score | 0.407673 (rank : 78) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
ARMX2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 81) | NC score | 0.019376 (rank : 108) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
FANCM_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 82) | NC score | 0.404540 (rank : 79) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
CC026_MOUSE
|
||||||
| θ value | 0.163984 (rank : 83) | NC score | 0.147605 (rank : 89) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9CZT6, Q9CY77 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26 homolog. | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 84) | NC score | 0.000664 (rank : 177) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
CD2L1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 85) | NC score | 0.000456 (rank : 178) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
GP149_MOUSE
|
||||||
| θ value | 0.21417 (rank : 86) | NC score | 0.031580 (rank : 97) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UVY1, Q3TMB1, Q80T52, Q8BXA3, Q8BZC0 | Gene names | Gpr149, Pgr10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 149 (G-protein coupled receptor PGR10). | |||||
|
IFIH1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 87) | NC score | 0.291840 (rank : 84) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
APTX_MOUSE
|
||||||
| θ value | 0.279714 (rank : 88) | NC score | 0.023730 (rank : 102) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TQC5, Q8BPA7, Q8C2B5, Q8K3D1, Q9CQ59 | Gene names | Aptx | |||
|
Domain Architecture |

|
|||||
| Description | Aprataxin homolog (Forkhead-associated domain histidine triad-like protein) (FHA-HIT). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 89) | NC score | 0.000034 (rank : 180) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
MP2K7_HUMAN
|
||||||
| θ value | 0.47712 (rank : 90) | NC score | -0.000278 (rank : 181) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14733, O14648, O14816, O60452, O60453 | Gene names | MAP2K7, JNKK2, MKK7, PRKMK7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity mitogen-activated protein kinase kinase 7 (EC 2.7.12.2) (MAP kinase kinase 7) (MAPKK 7) (MAPK/ERK kinase 7) (JNK-activating kinase 2) (c-Jun N-terminal kinase kinase 2) (JNK kinase 2) (JNKK 2). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 91) | NC score | 0.002995 (rank : 169) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
CCD41_HUMAN
|
||||||
| θ value | 0.62314 (rank : 92) | NC score | 0.004482 (rank : 166) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
IFIH1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 93) | NC score | 0.302618 (rank : 82) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
NCKX1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 94) | NC score | 0.015616 (rank : 119) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.62314 (rank : 95) | NC score | 0.012796 (rank : 127) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TMCO3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 96) | NC score | 0.016490 (rank : 116) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6UWJ1, Q5JSB1, Q6NUN1, Q8NG29, Q8TCI6, Q96EA6, Q9NWT2 | Gene names | TMCO3, C13orf11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domain-containing protein 3 precursor (Putative LAG1-interacting protein). | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 97) | NC score | 0.012311 (rank : 131) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 98) | NC score | 0.012441 (rank : 128) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
GTSE1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 99) | NC score | 0.017971 (rank : 112) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
PALM_MOUSE
|
||||||
| θ value | 0.813845 (rank : 100) | NC score | 0.021184 (rank : 106) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.813845 (rank : 101) | NC score | 0.007782 (rank : 156) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TAI12_MOUSE
|
||||||
| θ value | 0.813845 (rank : 102) | NC score | 0.017261 (rank : 114) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGQ2, Q3UIH1 | Gene names | Taip12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 103) | NC score | 0.011607 (rank : 133) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
GP149_HUMAN
|
||||||
| θ value | 1.06291 (rank : 104) | NC score | 0.027558 (rank : 99) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86SP6 | Gene names | GPR149, PGR10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 149 (G-protein coupled receptor PGR10). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 1.06291 (rank : 105) | NC score | 0.042207 (rank : 94) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 1.06291 (rank : 106) | NC score | 0.018706 (rank : 110) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
T22D3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 107) | NC score | 0.015060 (rank : 121) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99576, Q6FIH6, Q8NAI1, Q8WVB9, Q9UBN5, Q9UG13 | Gene names | TSC22D3, DSIPI, GILZ | |||
|
Domain Architecture |
|
|||||
| Description | TSC22 domain family protein 3 (Glucocorticoid-induced leucine zipper protein) (Delta sleep-inducing peptide immunoreactor) (DSIP- immunoreactive peptide) (Protein DIP) (hDIP) (TSC-22-like protein) (TSC-22R). | |||||
|
CHD8_HUMAN
|
||||||
| θ value | 1.38821 (rank : 108) | NC score | 0.013090 (rank : 126) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
CN092_MOUSE
|
||||||
| θ value | 1.38821 (rank : 109) | NC score | 0.010986 (rank : 137) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 1.38821 (rank : 110) | NC score | 0.011255 (rank : 135) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
STK33_HUMAN
|
||||||
| θ value | 1.38821 (rank : 111) | NC score | -0.004346 (rank : 191) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BYT3, Q658S6, Q8NEF5 | Gene names | STK33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 33 (EC 2.7.11.1). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 112) | NC score | 0.023519 (rank : 103) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
ABTAP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 113) | NC score | 0.011854 (rank : 132) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H501, Q86X92, Q9H9Q5, Q9HA35, Q9NX93, Q9P1S6 | Gene names | ABTAP, C20orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 114) | NC score | 0.010072 (rank : 141) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
MINT_MOUSE
|
||||||
| θ value | 1.81305 (rank : 115) | NC score | 0.019584 (rank : 107) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 1.81305 (rank : 116) | NC score | 0.044000 (rank : 93) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
TACC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 117) | NC score | 0.005752 (rank : 162) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 118) | NC score | 0.013133 (rank : 125) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
UBP10_MOUSE
|
||||||
| θ value | 1.81305 (rank : 119) | NC score | 0.009318 (rank : 144) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
AKAP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 120) | NC score | 0.011229 (rank : 136) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
ARX_HUMAN
|
||||||
| θ value | 2.36792 (rank : 121) | NC score | -0.000486 (rank : 182) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96QS3 | Gene names | ARX | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein ARX (Aristaless-related homeobox). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 2.36792 (rank : 122) | NC score | 0.008102 (rank : 152) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
ESCO1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 123) | NC score | 0.023330 (rank : 104) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 2.36792 (rank : 124) | NC score | 0.008061 (rank : 153) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RBBP8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 125) | NC score | 0.016052 (rank : 118) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99708, O75371 | Gene names | RBBP8, CTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 8 (RBBP-8) (CtBP-interacting protein) (CtIP) (Retinoblastoma-interacting protein and myosin-like) (RIM). | |||||
|
ST5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 126) | NC score | 0.008560 (rank : 148) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q924W7, Q78H54, Q8K2P3, Q924W8 | Gene names | St5, Dennd2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (DENN domain-containing protein 2B). | |||||
|
TRPS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 127) | NC score | 0.006326 (rank : 160) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
AFF2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 128) | NC score | 0.010028 (rank : 143) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
AP3B1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 129) | NC score | 0.007594 (rank : 158) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 130) | NC score | 0.010428 (rank : 139) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 131) | NC score | 0.014233 (rank : 123) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SIAL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 132) | NC score | 0.017722 (rank : 113) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61711, Q61363 | Gene names | Ibsp | |||
|
Domain Architecture |
|
|||||
| Description | Bone sialoprotein-2 precursor (Bone sialoprotein II) (BSP II) (Cell- binding sialoprotein) (Integrin-binding sialoprotein). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 133) | NC score | 0.007614 (rank : 157) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
TAI12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 134) | NC score | 0.014513 (rank : 122) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H175 | Gene names | TAIP12, C12orf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
TISD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 135) | NC score | 0.008772 (rank : 147) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47974, Q9BSJ3 | Gene names | ZFP36L2, BRF2, ERF2, TIS11D | |||
|
Domain Architecture |
|
|||||
| Description | Butyrate response factor 2 (TIS11D protein) (EGF-response factor 2) (ERF-2). | |||||
|
U520_HUMAN
|
||||||
| θ value | 3.0926 (rank : 136) | NC score | 0.131961 (rank : 90) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75643, O94884, Q6PX59, Q96IF2, Q9H7S0 | Gene names | ASCC3L1, HELIC2, KIAA0788 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U5 small nuclear ribonucleoprotein 200 kDa helicase (EC 3.6.1.-) (U5 snRNP-specific 200 kDa protein) (U5-200KD) (Activating signal cointegrator 1 complex subunit 3-like 1) (BRR2 homolog). | |||||
|
BRD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 137) | NC score | 0.005965 (rank : 161) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
JHD3C_MOUSE
|
||||||
| θ value | 4.03905 (rank : 138) | NC score | 0.003704 (rank : 167) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VCD7, Q3UNP7, Q69ZZ5, Q8BUY6, Q8BWA1 | Gene names | Jmjd2c, Jhdm3c, Kiaa0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C). | |||||
|
MCF2L_MOUSE
|
||||||
| θ value | 4.03905 (rank : 139) | NC score | 0.002918 (rank : 170) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64096 | Gene names | Mcf2l, Dbs | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide exchange factor DBS (DBL's big sister) (MCF2- transforming sequence-like protein). | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 140) | NC score | -0.001561 (rank : 185) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
PLCB2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 141) | NC score | 0.008514 (rank : 149) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q00722 | Gene names | PLCB2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-2) (PLC-beta-2). | |||||
|
TCF8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 142) | NC score | -0.005772 (rank : 194) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 4.03905 (rank : 143) | NC score | 0.018132 (rank : 111) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
ZBT17_MOUSE
|
||||||
| θ value | 4.03905 (rank : 144) | NC score | -0.007317 (rank : 196) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60821, Q60699 | Gene names | Zbtb17, Zfp100, Znf151 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Zinc finger protein 100) (Zfp-100) (Polyomavirus late initiator promoter-binding protein) (LP-1) (Zinc finger protein Z13). | |||||
|
ARX_MOUSE
|
||||||
| θ value | 5.27518 (rank : 145) | NC score | -0.001046 (rank : 184) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35085, Q9QYT4 | Gene names | Arx | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein ARX (Aristaless-related homeobox). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 146) | NC score | 0.021859 (rank : 105) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
CCDC4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 147) | NC score | 0.012400 (rank : 129) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZU67, Q58A26, Q58A27 | Gene names | CCDC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 4. | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 148) | NC score | 0.008145 (rank : 151) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 149) | NC score | 0.016768 (rank : 115) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
GSLG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 150) | NC score | 0.011515 (rank : 134) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92896, Q13221 | Gene names | GLG1, CFR1, ESL1, MG160 | |||
|
Domain Architecture |
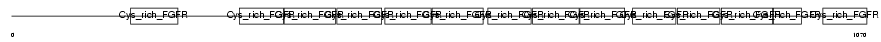
|
|||||
| Description | Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Cysteine-rich fibroblast growth factor receptor) (CFR-1). | |||||
|
MK15_MOUSE
|
||||||
| θ value | 5.27518 (rank : 151) | NC score | -0.003339 (rank : 190) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80Y86 | Gene names | Mapk15, Erk7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 7). | |||||
|
MO4L2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 152) | NC score | 0.013761 (rank : 124) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R0Q4, Q3UL62, Q6DIB1, Q6ZQK7, Q8C201, Q8C6C2 | Gene names | Morf4l2, Kiaa0026, Sid393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (Sid 393). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 153) | NC score | 0.024441 (rank : 101) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 154) | NC score | 0.008194 (rank : 150) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
ST5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 155) | NC score | 0.008033 (rank : 154) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78524, P78523, Q16492, Q7KYY2, Q7KZ12, Q8NE12, Q9BQQ6 | Gene names | ST5, DENND2B, HTS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (HeLa tumor suppression 1) (DENN domain-containing protein 2B). | |||||
|
ZBTB3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 156) | NC score | -0.004737 (rank : 193) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H5J0 | Gene names | ZBTB3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 3. | |||||
|
ZN142_HUMAN
|
||||||
| θ value | 5.27518 (rank : 157) | NC score | -0.006429 (rank : 195) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
ASTL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 158) | NC score | 0.005390 (rank : 163) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6HA08 | Gene names | ASTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
CG1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 159) | NC score | 0.016097 (rank : 117) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 6.88961 (rank : 160) | NC score | 0.018964 (rank : 109) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
DZIP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 161) | NC score | 0.002697 (rank : 173) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BMD2, Q6ZQ10, Q9CRK5 | Gene names | Dzip1, Kiaa0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1 homolog). | |||||
|
GSLG1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 162) | NC score | 0.010860 (rank : 138) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61543, Q9QZ40 | Gene names | Glg1, Esl1, Mg160, Selel | |||
|
Domain Architecture |
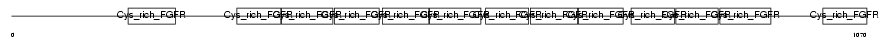
|
|||||
| Description | Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Selel). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 163) | NC score | 0.036268 (rank : 95) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LGP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 164) | NC score | 0.288408 (rank : 85) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
MADCA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 165) | NC score | 0.008900 (rank : 146) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 166) | NC score | 0.015133 (rank : 120) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MYPT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 167) | NC score | 0.000895 (rank : 175) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
NCA11_MOUSE
|
||||||
| θ value | 6.88961 (rank : 168) | NC score | -0.001860 (rank : 186) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 169) | NC score | 0.026728 (rank : 100) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
SEL1L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 170) | NC score | 0.004822 (rank : 164) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
SEPT9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 171) | NC score | 0.003294 (rank : 168) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80UG5, Q3URP2, Q80TM7, Q9QYX9 | Gene names | Sept9, Kiaa0991, Sint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (SL3-3 integration site 1 protein). | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | -0.002275 (rank : 187) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.012339 (rank : 130) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.010066 (rank : 142) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
CCD96_MOUSE
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | 0.002773 (rank : 171) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
CPSF7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 176) | NC score | 0.004604 (rank : 165) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N684, Q7Z3H9, Q9H025, Q9H9V1 | Gene names | CPSF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7 (Cleavage and polyadenylation specificity factor 59 kDa subunit) (CPSF 59 kDa subunit) (Pre-mRNA cleavage factor Im 59 kDa subunit). | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 177) | NC score | -0.003157 (rank : 189) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 178) | NC score | 0.007902 (rank : 155) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
EHMT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 179) | NC score | -0.000527 (rank : 183) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 180) | NC score | 0.029609 (rank : 98) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 181) | NC score | 0.000665 (rank : 176) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYLK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 182) | NC score | -0.004578 (rank : 192) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H1R3, Q96I84 | Gene names | MYLK2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain kinase 2, skeletal/cardiac muscle (EC 2.7.11.18) (MLCK2). | |||||
|
NEBL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 183) | NC score | 0.002090 (rank : 174) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O76041, Q9UIC4 | Gene names | NEBL | |||
|
Domain Architecture |

|
|||||
| Description | Nebulette (Actin-binding Z-disk protein). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 8.99809 (rank : 184) | NC score | 0.006380 (rank : 159) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
NU214_HUMAN
|
||||||
| θ value | 8.99809 (rank : 185) | NC score | 0.010327 (rank : 140) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
OS9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 186) | NC score | 0.009152 (rank : 145) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13438, O00579 | Gene names | OS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein OS-9 precursor (Amplified in osteosarcoma 9). | |||||
|
PYGM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 187) | NC score | 0.002716 (rank : 172) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUB3 | Gene names | Pygm | |||
|
Domain Architecture |

|
|||||
| Description | Glycogen phosphorylase, muscle form (EC 2.4.1.1) (Myophosphorylase). | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 188) | NC score | 0.430230 (rank : 75) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
TXLNG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 189) | NC score | 0.000142 (rank : 179) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHN1, Q8BP11 | Gene names | Txlng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin. | |||||
|
WEE1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 190) | NC score | -0.002438 (rank : 188) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P0C1S8 | Gene names | WEE1B, WEE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wee1-like protein kinase 1B (EC 2.7.10.2) (Wee1B kinase). | |||||
|
CC026_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.162077 (rank : 87) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BQ75 | Gene names | C3orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26. | |||||
|
DDX58_HUMAN
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.153440 (rank : 88) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
DDX58_MOUSE
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.184295 (rank : 86) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
HELC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.089487 (rank : 91) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
LGP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.296520 (rank : 83) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
SMAL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 196) | NC score | 0.073053 (rank : 92) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
DDX24_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 190 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX24_MOUSE
|
||||||
| NC score | 0.992652 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX47_MOUSE
|
||||||
| NC score | 0.949864 (rank : 3) | θ value | 8.30557e-46 (rank : 4) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX47_HUMAN
|
||||||
| NC score | 0.949644 (rank : 4) | θ value | 3.72821e-46 (rank : 3) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX49_MOUSE
|
||||||
| NC score | 0.945605 (rank : 5) | θ value | 1.6774e-38 (rank : 6) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX49_HUMAN
|
||||||
| NC score | 0.943132 (rank : 6) | θ value | 1.79215e-40 (rank : 5) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX27_MOUSE
|
||||||
| NC score | 0.934666 (rank : 7) | θ value | 9.52487e-34 (rank : 9) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX18_HUMAN
|
||||||
| NC score | 0.933609 (rank : 8) | θ value | 1.46948e-34 (rank : 7) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
DDX18_MOUSE
|
||||||
| NC score | 0.933474 (rank : 9) | θ value | 2.50655e-34 (rank : 8) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
DDX55_MOUSE
|
||||||
| NC score | 0.933048 (rank : 10) | θ value | 1.4233e-29 (rank : 20) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX41_HUMAN
|
||||||
| NC score | 0.932960 (rank : 11) | θ value | 2.77131e-33 (rank : 10) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
DDX27_HUMAN
|
||||||
| NC score | 0.932956 (rank : 12) | θ value | 6.17384e-33 (rank : 14) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX41_MOUSE
|
||||||
| NC score | 0.932641 (rank : 13) | θ value | 2.77131e-33 (rank : 11) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
DDX54_MOUSE
|
||||||
| NC score | 0.931930 (rank : 14) | θ value | 1.52067e-31 (rank : 15) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
DDX55_HUMAN
|
||||||
| NC score | 0.931458 (rank : 15) | θ value | 1.08979e-29 (rank : 19) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX31_HUMAN
|
||||||
| NC score | 0.930129 (rank : 16) | θ value | 1.92365e-26 (rank : 32) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
DDX28_MOUSE
|
||||||
| NC score | 0.928753 (rank : 17) | θ value | 1.24688e-25 (rank : 38) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX54_HUMAN
|
||||||
| NC score | 0.927540 (rank : 18) | θ value | 8.3442e-30 (rank : 18) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
DDX28_HUMAN
|
||||||
| NC score | 0.926474 (rank : 19) | θ value | 1.24688e-25 (rank : 37) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX17_MOUSE
|
||||||
| NC score | 0.926298 (rank : 20) | θ value | 6.17384e-33 (rank : 13) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
DDX17_HUMAN
|
||||||
| NC score | 0.926165 (rank : 21) | θ value | 6.17384e-33 (rank : 12) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
DDX10_HUMAN
|
||||||
| NC score | 0.925860 (rank : 22) | θ value | 5.77852e-31 (rank : 16) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX10_MOUSE
|
||||||
| NC score | 0.925287 (rank : 23) | θ value | 4.89182e-30 (rank : 17) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX43_HUMAN
|
||||||
| NC score | 0.924561 (rank : 24) | θ value | 3.87602e-27 (rank : 27) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
DDX6_HUMAN
|
||||||
| NC score | 0.923597 (rank : 25) | θ value | 5.23862e-24 (rank : 48) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
DDX6_MOUSE
|
||||||
| NC score | 0.923585 (rank : 26) | θ value | 5.23862e-24 (rank : 49) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
DDX56_HUMAN
|
||||||
| NC score | 0.922389 (rank : 27) | θ value | 6.61148e-27 (rank : 29) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NY93, Q9H9I8 | Gene names | DDX56, DDX21, NOH61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase) (DEAD-box protein 21). | |||||
|
DDX56_MOUSE
|
||||||
| NC score | 0.921299 (rank : 28) | θ value | 1.12775e-26 (rank : 31) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
DDX5_HUMAN
|
||||||
| NC score | 0.919632 (rank : 29) | θ value | 5.40856e-29 (rank : 21) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
DDX5_MOUSE
|
||||||
| NC score | 0.919611 (rank : 30) | θ value | 5.40856e-29 (rank : 22) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
DDX48_HUMAN
|
||||||
| NC score | 0.918634 (rank : 31) | θ value | 5.97985e-28 (rank : 25) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX48_MOUSE
|
||||||
| NC score | 0.918482 (rank : 32) | θ value | 5.97985e-28 (rank : 26) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
DDX4_MOUSE
|
||||||
| NC score | 0.917514 (rank : 33) | θ value | 8.93572e-24 (rank : 50) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
DDX4_HUMAN
|
||||||
| NC score | 0.916946 (rank : 34) | θ value | 5.23862e-24 (rank : 46) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
DDX53_HUMAN
|
||||||
| NC score | 0.916856 (rank : 35) | θ value | 1.5242e-23 (rank : 53) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
DDX39_HUMAN
|
||||||
| NC score | 0.915274 (rank : 36) | θ value | 1.05554e-24 (rank : 43) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O00148, Q9BVP6, Q9H5W0 | Gene names | DDX39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39) (Nuclear RNA helicase URH49). | |||||
|
DDX39_MOUSE
|
||||||
| NC score | 0.914840 (rank : 37) | θ value | 1.80048e-24 (rank : 44) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8VDW0, Q3UJV4, Q8C2C2 | Gene names | Ddx39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39). | |||||
|
DDX51_MOUSE
|
||||||
| NC score | 0.914404 (rank : 38) | θ value | 4.43474e-23 (rank : 54) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
UAP56_HUMAN
|
||||||
| NC score | 0.913802 (rank : 39) | θ value | 8.93572e-24 (rank : 51) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q13838, O43496, Q5RJ64, Q5RJ66, Q5ST94, Q5STB4, Q5STB5, Q5STB7, Q5STU4, Q5STU5, Q5STU6, Q5STU8, Q71V76 | Gene names | BAT1, UAP56 | |||
|
Domain Architecture |

|
|||||
| Description | Spliceosome RNA helicase BAT1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (ATP-dependent RNA helicase p47) (HLA-B-associated transcript-1). | |||||
|
UAP56_MOUSE
|
||||||
| NC score | 0.913732 (rank : 40) | θ value | 8.93572e-24 (rank : 52) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9Z1N5, Q8HW97 | Gene names | Bat1, Bat1a, Uap56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spliceosome RNA helicase Bat1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (HLA-B-associated transcript 1). | |||||
|
DDX52_HUMAN
|
||||||
| NC score | 0.913060 (rank : 41) | θ value | 2.13179e-17 (rank : 62) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
IF4A1_HUMAN
|
||||||
| NC score | 0.912849 (rank : 42) | θ value | 4.73814e-25 (rank : 39) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| NC score | 0.912849 (rank : 43) | θ value | 4.73814e-25 (rank : 40) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
DDX52_MOUSE
|
||||||
| NC score | 0.912606 (rank : 44) | θ value | 2.13179e-17 (rank : 63) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8K301, Q8BV29 | Gene names | Ddx52, Rok1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
IF4A2_HUMAN
|
||||||
| NC score | 0.912483 (rank : 45) | θ value | 4.73814e-25 (rank : 41) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| NC score | 0.912483 (rank : 46) | θ value | 4.73814e-25 (rank : 42) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
DDX3Y_MOUSE
|
||||||
| NC score | 0.911440 (rank : 47) | θ value | 1.12775e-26 (rank : 30) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
DDX50_MOUSE
|
||||||
| NC score | 0.911056 (rank : 48) | θ value | 4.01107e-24 (rank : 45) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX50_HUMAN
|
||||||
| NC score | 0.910536 (rank : 49) | θ value | 5.23862e-24 (rank : 47) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
PL10_MOUSE
|
||||||
| NC score | 0.910320 (rank : 50) | θ value | 4.28545e-26 (rank : 35) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
DDX3X_HUMAN
|
||||||
| NC score | 0.910277 (rank : 51) | θ value | 1.92365e-26 (rank : 33) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX3X_MOUSE
|
||||||
| NC score | 0.909567 (rank : 52) | θ value | 4.28545e-26 (rank : 34) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
DDX3Y_HUMAN
|
||||||
| NC score | 0.907734 (rank : 53) | θ value | 9.54697e-26 (rank : 36) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
DDX46_HUMAN
|
||||||
| NC score | 0.907071 (rank : 54) | θ value | 5.97985e-28 (rank : 23) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX46_MOUSE
|
||||||
| NC score | 0.905425 (rank : 55) | θ value | 5.97985e-28 (rank : 24) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
DDX23_HUMAN
|
||||||
| NC score | 0.900346 (rank : 56) | θ value | 6.61148e-27 (rank : 28) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.899497 (rank : 57) | θ value | 5.79196e-23 (rank : 55) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX20_HUMAN
|
||||||
| NC score | 0.898933 (rank : 58) | θ value | 6.62687e-19 (rank : 61) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
DDX51_HUMAN
|
||||||
| NC score | 0.897591 (rank : 59) | θ value | 1.86321e-21 (rank : 59) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX20_MOUSE
|
||||||
| NC score | 0.896185 (rank : 60) | θ value | 3.88503e-19 (rank : 60) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.892264 (rank : 61) | θ value | 8.36355e-22 (rank : 57) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DD19A_HUMAN
|
||||||
| NC score | 0.881676 (rank : 62) | θ value | 2.35696e-16 (rank : 64) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19B_HUMAN
|
||||||
| NC score | 0.880638 (rank : 63) | θ value | 4.02038e-16 (rank : 65) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
DD19A_MOUSE
|
||||||
| NC score | 0.879955 (rank : 64) | θ value | 1.52774e-15 (rank : 66) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DDX25_HUMAN
|
||||||
| NC score | 0.878419 (rank : 65) | θ value | 9.26847e-13 (rank : 68) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX25_MOUSE
|
||||||
| NC score | 0.877940 (rank : 66) | θ value | 3.18553e-13 (rank : 67) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX1_MOUSE
|
||||||
| NC score | 0.870999 (rank : 67) | θ value | 3.75424e-22 (rank : 56) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
DDX1_HUMAN
|
||||||
| NC score | 0.869761 (rank : 68) | θ value | 1.09232e-21 (rank : 58) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
RECQ4_HUMAN
|
||||||
| NC score | 0.526881 (rank : 69) | θ value | 0.0193708 (rank : 78) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
RECQ4_MOUSE
|
||||||
| NC score | 0.510439 (rank : 70) | θ value | 0.0113563 (rank : 77) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
BLM_MOUSE
|
||||||
| NC score | 0.484691 (rank : 71) | θ value | 0.00665767 (rank : 75) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O88700, O88198 | Gene names | Blm | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein homolog (EC 3.6.1.-) (mBLM). | |||||
|
BLM_HUMAN
|
||||||
| NC score | 0.473534 (rank : 72) | θ value | 0.00509761 (rank : 74) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
WRN_HUMAN
|
||||||
| NC score | 0.468722 (rank : 73) | θ value | 8.40245e-06 (rank : 69) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
RECQ1_MOUSE
|
||||||
| NC score | 0.434389 (rank : 74) | θ value | 0.000121331 (rank : 70) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.430230 (rank : 75) | θ value | 8.99809 (rank : 188) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
RECQ1_HUMAN
|
||||||
| NC score | 0.429340 (rank : 76) | θ value | 0.00869519 (rank : 76) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P46063 | Gene names | RECQL, RECQL1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
WRN_MOUSE
|
||||||
| NC score | 0.429309 (rank : 77) | θ value | 0.00102713 (rank : 73) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
FANCM_HUMAN
|
||||||
| NC score | 0.407673 (rank : 78) | θ value | 0.0563607 (rank : 80) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
FANCM_MOUSE
|
||||||
| NC score | 0.404540 (rank : 79) | θ value | 0.0736092 (rank : 82) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
DICER_HUMAN
|
||||||
| NC score | 0.402072 (rank : 80) | θ value | 0.00102713 (rank : 71) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
DICER_MOUSE
|
||||||
| NC score | 0.401518 (rank : 81) | θ value | 0.00102713 (rank : 72) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
IFIH1_HUMAN
|
||||||
| NC score | 0.302618 (rank : 82) | θ value | 0.62314 (rank : 93) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
LGP2_MOUSE
|
||||||
| NC score | 0.296520 (rank : 83) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
IFIH1_MOUSE
|
||||||
| NC score | 0.291840 (rank : 84) | θ value | 0.21417 (rank : 87) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
LGP2_HUMAN
|
||||||
| NC score | 0.288408 (rank : 85) | θ value | 6.88961 (rank : 164) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
DDX58_MOUSE
|
||||||
| NC score | 0.184295 (rank : 86) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
CC026_HUMAN
|
||||||
| NC score | 0.162077 (rank : 87) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BQ75 | Gene names | C3orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26. | |||||
|
DDX58_HUMAN
|
||||||
| NC score | 0.153440 (rank : 88) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
CC026_MOUSE
|
||||||
| NC score | 0.147605 (rank : 89) | θ value | 0.163984 (rank : 83) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9CZT6, Q9CY77 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26 homolog. | |||||
|
U520_HUMAN
|
||||||
| NC score | 0.131961 (rank : 90) | θ value | 3.0926 (rank : 136) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75643, O94884, Q6PX59, Q96IF2, Q9H7S0 | Gene names | ASCC3L1, HELIC2, KIAA0788 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U5 small nuclear ribonucleoprotein 200 kDa helicase (EC 3.6.1.-) (U5 snRNP-specific 200 kDa protein) (U5-200KD) (Activating signal cointegrator 1 complex subunit 3-like 1) (BRR2 homolog). | |||||
|
HELC1_HUMAN
|
||||||
| NC score | 0.089487 (rank : 91) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
SMAL1_HUMAN
|
||||||
| NC score | 0.073053 (rank : 92) | θ value | θ > 10 (rank : 196) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.044000 (rank : 93) | θ value | 1.81305 (rank : 116) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.042207 (rank : 94) | θ value | 1.06291 (rank : 105) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.036268 (rank : 95) | θ value | 6.88961 (rank : 163) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
TTC16_HUMAN
|
||||||
| NC score | 0.034128 (rank : 96) | θ value | 0.0252991 (rank : 79) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
GP149_MOUSE
|
||||||
| NC score | 0.031580 (rank : 97) | θ value | 0.21417 (rank : 86) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UVY1, Q3TMB1, Q80T52, Q8BXA3, Q8BZC0 | Gene names | Gpr149, Pgr10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 149 (G-protein coupled receptor PGR10). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.029609 (rank : 98) | θ value | 8.99809 (rank : 180) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
GP149_HUMAN
|
||||||
| NC score | 0.027558 (rank : 99) | θ value | 1.06291 (rank : 104) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86SP6 | Gene names | GPR149, PGR10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 149 (G-protein coupled receptor PGR10). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.026728 (rank : 100) | θ value | 6.88961 (rank : 169) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.024441 (rank : 101) | θ value | 5.27518 (rank : 153) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
APTX_MOUSE
|
||||||
| NC score | 0.023730 (rank : 102) | θ value | 0.279714 (rank : 88) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TQC5, Q8BPA7, Q8C2B5, Q8K3D1, Q9CQ59 | Gene names | Aptx | |||
|
Domain Architecture |

|
|||||
| Description | Aprataxin homolog (Forkhead-associated domain histidine triad-like protein) (FHA-HIT). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.023519 (rank : 103) | θ value | 1.38821 (rank : 112) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
ESCO1_HUMAN
|
||||||
| NC score | 0.023330 (rank : 104) | θ value | 2.36792 (rank : 123) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.021859 (rank : 105) | θ value | 5.27518 (rank : 146) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
PALM_MOUSE
|
||||||
| NC score | 0.021184 (rank : 106) | θ value | 0.813845 (rank : 100) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.019584 (rank : 107) | θ value | 1.81305 (rank : 115) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
ARMX2_HUMAN
|
||||||
| NC score | 0.019376 (rank : 108) | θ value | 0.0736092 (rank : 81) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.018964 (rank : 109) | θ value | 6.88961 (rank : 160) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.018706 (rank : 110) | θ value | 1.06291 (rank : 106) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.018132 (rank : 111) | θ value | 4.03905 (rank : 143) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
GTSE1_MOUSE
|
||||||
| NC score | 0.017971 (rank : 112) | θ value | 0.813845 (rank : 99) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
SIAL_MOUSE
|
||||||
| NC score | 0.017722 (rank : 113) | θ value | 3.0926 (rank : 132) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61711, Q61363 | Gene names | Ibsp | |||
|
Domain Architecture |

|
|||||
| Description | Bone sialoprotein-2 precursor (Bone sialoprotein II) (BSP II) (Cell- binding sialoprotein) (Integrin-binding sialoprotein). | |||||
|
TAI12_MOUSE
|
||||||
| NC score | 0.017261 (rank : 114) | θ value | 0.813845 (rank : 102) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGQ2, Q3UIH1 | Gene names | Taip12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.016768 (rank : 115) | θ value | 5.27518 (rank : 149) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
TMCO3_HUMAN
|
||||||
| NC score | 0.016490 (rank : 116) | θ value | 0.62314 (rank : 96) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6UWJ1, Q5JSB1, Q6NUN1, Q8NG29, Q8TCI6, Q96EA6, Q9NWT2 | Gene names | TMCO3, C13orf11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domain-containing protein 3 precursor (Putative LAG1-interacting protein). | |||||
|
CG1_HUMAN
|
||||||
| NC score | 0.016097 (rank : 117) | θ value | 6.88961 (rank : 159) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
RBBP8_HUMAN
|
||||||
| NC score | 0.016052 (rank : 118) | θ value | 2.36792 (rank : 125) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99708, O75371 | Gene names | RBBP8, CTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 8 (RBBP-8) (CtBP-interacting protein) (CtIP) (Retinoblastoma-interacting protein and myosin-like) (RIM). | |||||
|
NCKX1_HUMAN
|
||||||
| NC score | 0.015616 (rank : 119) | θ value | 0.62314 (rank : 94) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.015133 (rank : 120) | θ value | 6.88961 (rank : 166) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
T22D3_HUMAN
|
||||||
| NC score | 0.015060 (rank : 121) | θ value | 1.06291 (rank : 107) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99576, Q6FIH6, Q8NAI1, Q8WVB9, Q9UBN5, Q9UG13 | Gene names | TSC22D3, DSIPI, GILZ | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 3 (Glucocorticoid-induced leucine zipper protein) (Delta sleep-inducing peptide immunoreactor) (DSIP- immunoreactive peptide) (Protein DIP) (hDIP) (TSC-22-like protein) (TSC-22R). | |||||
|
TAI12_HUMAN
|
||||||
| NC score | 0.014513 (rank : 122) | θ value | 3.0926 (rank : 134) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H175 | Gene names | TAIP12, C12orf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.014233 (rank : 123) | θ value | 3.0926 (rank : 131) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
MO4L2_MOUSE
|
||||||
| NC score | 0.013761 (rank : 124) | θ value | 5.27518 (rank : 152) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R0Q4, Q3UL62, Q6DIB1, Q6ZQK7, Q8C201, Q8C6C2 | Gene names | Morf4l2, Kiaa0026, Sid393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (Sid 393). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.013133 (rank : 125) | θ value | 1.81305 (rank : 118) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
CHD8_HUMAN
|
||||||
| NC score | 0.013090 (rank : 126) | θ value | 1.38821 (rank : 108) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.012796 (rank : 127) | θ value | 0.62314 (rank : 95) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.012441 (rank : 128) | θ value | 0.813845 (rank : 98) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
CCDC4_HUMAN
|
||||||
| NC score | 0.012400 (rank : 129) | θ value | 5.27518 (rank : 147) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZU67, Q58A26, Q58A27 | Gene names | CCDC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 4. | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.012339 (rank : 130) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.012311 (rank : 131) | θ value | 0.813845 (rank : 97) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ABTAP_HUMAN
|
||||||
| NC score | 0.011854 (rank : 132) | θ value | 1.81305 (rank : 113) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H501, Q86X92, Q9H9Q5, Q9HA35, Q9NX93, Q9P1S6 | Gene names | ABTAP, C20orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.011607 (rank : 133) | θ value | 1.06291 (rank : 103) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
GSLG1_HUMAN
|
||||||
| NC score | 0.011515 (rank : 134) | θ value | 5.27518 (rank : 150) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92896, Q13221 | Gene names | GLG1, CFR1, ESL1, MG160 | |||
|
Domain Architecture |
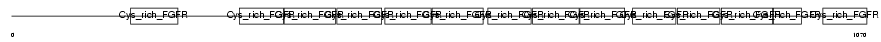
|
|||||
| Description | Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Cysteine-rich fibroblast growth factor receptor) (CFR-1). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.011255 (rank : 135) | θ value | 1.38821 (rank : 110) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
AKAP1_HUMAN
|
||||||
| NC score | 0.011229 (rank : 136) | θ value | 2.36792 (rank : 120) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
CN092_MOUSE
|
||||||
| NC score | 0.010986 (rank : 137) | θ value | 1.38821 (rank : 109) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
GSLG1_MOUSE
|
||||||
| NC score | 0.010860 (rank : 138) | θ value | 6.88961 (rank : 162) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61543, Q9QZ40 | Gene names | Glg1, Esl1, Mg160, Selel | |||
|
Domain Architecture |
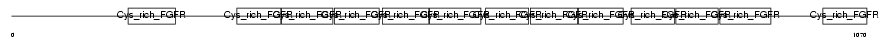
|
|||||
| Description | Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Selel). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.010428 (rank : 139) | θ value | 3.0926 (rank : 130) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.010327 (rank : 140) | θ value | 8.99809 (rank : 185) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.010072 (rank : 141) | θ value | 1.81305 (rank : 114) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.010066 (rank : 142) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
AFF2_HUMAN
|
||||||
| NC score | 0.010028 (rank : 143) | θ value | 3.0926 (rank : 128) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
UBP10_MOUSE
|
||||||
| NC score | 0.009318 (rank : 144) | θ value | 1.81305 (rank : 119) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
OS9_HUMAN
|
||||||
| NC score | 0.009152 (rank : 145) | θ value | 8.99809 (rank : 186) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13438, O00579 | Gene names | OS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein OS-9 precursor (Amplified in osteosarcoma 9). | |||||
|
MADCA_HUMAN
|
||||||
| NC score | 0.008900 (rank : 146) | θ value | 6.88961 (rank : 165) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
TISD_HUMAN
|
||||||
| NC score | 0.008772 (rank : 147) | θ value | 3.0926 (rank : 135) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47974, Q9BSJ3 | Gene names | ZFP36L2, BRF2, ERF2, TIS11D | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 2 (TIS11D protein) (EGF-response factor 2) (ERF-2). | |||||
|
ST5_MOUSE
|
||||||
| NC score | 0.008560 (rank : 148) | θ value | 2.36792 (rank : 126) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q924W7, Q78H54, Q8K2P3, Q924W8 | Gene names | St5, Dennd2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (DENN domain-containing protein 2B). | |||||
|
PLCB2_HUMAN
|
||||||
| NC score | 0.008514 (rank : 149) | θ value | 4.03905 (rank : 141) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q00722 | Gene names | PLCB2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-2) (PLC-beta-2). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.008194 (rank : 150) | θ value | 5.27518 (rank : 154) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.008145 (rank : 151) | θ value | 5.27518 (rank : 148) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.008102 (rank : 152) | θ value | 2.36792 (rank : 122) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.008061 (rank : 153) | θ value | 2.36792 (rank : 124) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ST5_HUMAN
|
||||||
| NC score | 0.008033 (rank : 154) | θ value | 5.27518 (rank : 155) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78524, P78523, Q16492, Q7KYY2, Q7KZ12, Q8NE12, Q9BQQ6 | Gene names | ST5, DENND2B, HTS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (HeLa tumor suppression 1) (DENN domain-containing protein 2B). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.007902 (rank : 155) | θ value | 8.99809 (rank : 178) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.007782 (rank : 156) | θ value | 0.813845 (rank : 101) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.007614 (rank : 157) | θ value | 3.0926 (rank : 133) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
AP3B1_HUMAN
|
||||||
| NC score | 0.007594 (rank : 158) | θ value | 3.0926 (rank : 129) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.006380 (rank : 159) | θ value | 8.99809 (rank : 184) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
TRPS1_MOUSE
|
||||||
| NC score | 0.006326 (rank : 160) | θ value | 2.36792 (rank : 127) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
BRD2_HUMAN
|
||||||
| NC score | 0.005965 (rank : 161) | θ value | 4.03905 (rank : 137) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
TACC2_MOUSE
|
||||||
| NC score | 0.005752 (rank : 162) | θ value | 1.81305 (rank : 117) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
ASTL_HUMAN
|
||||||
| NC score | 0.005390 (rank : 163) | θ value | 6.88961 (rank : 158) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6HA08 | Gene names | ASTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
SEL1L_MOUSE
|
||||||
| NC score | 0.004822 (rank : 164) | θ value | 6.88961 (rank : 170) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
CPSF7_HUMAN
|
||||||
| NC score | 0.004604 (rank : 165) | θ value | 8.99809 (rank : 176) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N684, Q7Z3H9, Q9H025, Q9H9V1 | Gene names | CPSF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7 (Cleavage and polyadenylation specificity factor 59 kDa subunit) (CPSF 59 kDa subunit) (Pre-mRNA cleavage factor Im 59 kDa subunit). | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.004482 (rank : 166) | θ value | 0.62314 (rank : 92) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
JHD3C_MOUSE
|
||||||
| NC score | 0.003704 (rank : 167) | θ value | 4.03905 (rank : 138) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VCD7, Q3UNP7, Q69ZZ5, Q8BUY6, Q8BWA1 | Gene names | Jmjd2c, Jhdm3c, Kiaa0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C). | |||||
|
SEPT9_MOUSE
|
||||||
| NC score | 0.003294 (rank : 168) | θ value | 6.88961 (rank : 171) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80UG5, Q3URP2, Q80TM7, Q9QYX9 | Gene names | Sept9, Kiaa0991, Sint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (SL3-3 integration site 1 protein). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.002995 (rank : 169) | θ value | 0.62314 (rank : 91) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
MCF2L_MOUSE
|
||||||
| NC score | 0.002918 (rank : 170) | θ value | 4.03905 (rank : 139) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64096 | Gene names | Mcf2l, Dbs | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide exchange factor DBS (DBL's big sister) (MCF2- transforming sequence-like protein). | |||||
|
CCD96_MOUSE
|
||||||
| NC score | 0.002773 (rank : 171) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
PYGM_MOUSE
|
||||||
| NC score | 0.002716 (rank : 172) | θ value | 8.99809 (rank : 187) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUB3 | Gene names | Pygm | |||
|
Domain Architecture |

|
|||||
| Description | Glycogen phosphorylase, muscle form (EC 2.4.1.1) (Myophosphorylase). | |||||
|
DZIP1_MOUSE
|
||||||
| NC score | 0.002697 (rank : 173) | θ value | 6.88961 (rank : 161) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BMD2, Q6ZQ10, Q9CRK5 | Gene names | Dzip1, Kiaa0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1 homolog). | |||||
|
NEBL_HUMAN
|
||||||
| NC score | 0.002090 (rank : 174) | θ value | 8.99809 (rank : 183) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O76041, Q9UIC4 | Gene names | NEBL | |||
|
Domain Architecture |

|
|||||
| Description | Nebulette (Actin-binding Z-disk protein). | |||||
|
MYPT1_MOUSE
|
||||||
| NC score | 0.000895 (rank : 175) | θ value | 6.88961 (rank : 167) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.000665 (rank : 176) | θ value | 8.99809 (rank : 181) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.000664 (rank : 177) | θ value | 0.21417 (rank : 84) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
CD2L1_MOUSE
|
||||||
| NC score | 0.000456 (rank : 178) | θ value | 0.21417 (rank : 85) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
TXLNG_MOUSE
|
||||||
| NC score | 0.000142 (rank : 179) | θ value | 8.99809 (rank : 189) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHN1, Q8BP11 | Gene names | Txlng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin. | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.000034 (rank : 180) | θ value | 0.47712 (rank : 89) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
MP2K7_HUMAN
|
||||||
| NC score | -0.000278 (rank : 181) | θ value | 0.47712 (rank : 90) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14733, O14648, O14816, O60452, O60453 | Gene names | MAP2K7, JNKK2, MKK7, PRKMK7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity mitogen-activated protein kinase kinase 7 (EC 2.7.12.2) (MAP kinase kinase 7) (MAPKK 7) (MAPK/ERK kinase 7) (JNK-activating kinase 2) (c-Jun N-terminal kinase kinase 2) (JNK kinase 2) (JNKK 2). | |||||
|
ARX_HUMAN
|
||||||
| NC score | -0.000486 (rank : 182) | θ value | 2.36792 (rank : 121) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96QS3 | Gene names | ARX | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein ARX (Aristaless-related homeobox). | |||||
|
EHMT1_HUMAN
|
||||||
| NC score | -0.000527 (rank : 183) | θ value | 8.99809 (rank : 179) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
ARX_MOUSE
|
||||||
| NC score | -0.001046 (rank : 184) | θ value | 5.27518 (rank : 145) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35085, Q9QYT4 | Gene names | Arx | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein ARX (Aristaless-related homeobox). | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | -0.001561 (rank : 185) | θ value | 4.03905 (rank : 140) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
NCA11_MOUSE
|
||||||
| NC score | -0.001860 (rank : 186) | θ value | 6.88961 (rank : 168) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
ANR26_MOUSE
|
||||||
| NC score | -0.002275 (rank : 187) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
WEE1B_HUMAN
|
||||||
| NC score | -0.002438 (rank : 188) | θ value | 8.99809 (rank : 190) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P0C1S8 | Gene names | WEE1B, WEE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wee1-like protein kinase 1B (EC 2.7.10.2) (Wee1B kinase). | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | -0.003157 (rank : 189) | θ value | 8.99809 (rank : 177) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
MK15_MOUSE
|
||||||
| NC score | -0.003339 (rank : 190) | θ value | 5.27518 (rank : 151) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80Y86 | Gene names | Mapk15, Erk7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 7). | |||||
|
STK33_HUMAN
|
||||||
| NC score | -0.004346 (rank : 191) | θ value | 1.38821 (rank : 111) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BYT3, Q658S6, Q8NEF5 | Gene names | STK33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 33 (EC 2.7.11.1). | |||||
|
MYLK2_HUMAN
|
||||||
| NC score | -0.004578 (rank : 192) | θ value | 8.99809 (rank : 182) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H1R3, Q96I84 | Gene names | MYLK2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain kinase 2, skeletal/cardiac muscle (EC 2.7.11.18) (MLCK2). | |||||
|
ZBTB3_HUMAN
|
||||||
| NC score | -0.004737 (rank : 193) | θ value | 5.27518 (rank : 156) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H5J0 | Gene names | ZBTB3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 3. | |||||
|
TCF8_MOUSE
|
||||||
| NC score | -0.005772 (rank : 194) | θ value | 4.03905 (rank : 142) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
ZN142_HUMAN
|
||||||
| NC score | -0.006429 (rank : 195) | θ value | 5.27518 (rank : 157) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
ZBT17_MOUSE
|
||||||
| NC score | -0.007317 (rank : 196) | θ value | 4.03905 (rank : 144) | |||
| Query Neighborhood Hits | 190 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60821, Q60699 | Gene names | Zbtb17, Zfp100, Znf151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Zinc finger protein 100) (Zfp-100) (Polyomavirus late initiator promoter-binding protein) (LP-1) (Zinc finger protein Z13). | |||||