Please be patient as the page loads
|
GSLG1_MOUSE
|
||||||
| SwissProt Accessions | Q61543, Q9QZ40 | Gene names | Glg1, Esl1, Mg160, Selel | |||
|
Domain Architecture |
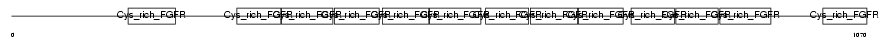
|
|||||
| Description | Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Selel). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GSLG1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.995172 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q92896, Q13221 | Gene names | GLG1, CFR1, ESL1, MG160 | |||
|
Domain Architecture |
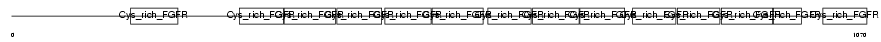
|
|||||
| Description | Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Cysteine-rich fibroblast growth factor receptor) (CFR-1). | |||||
|
GSLG1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61543, Q9QZ40 | Gene names | Glg1, Esl1, Mg160, Selel | |||
|
Domain Architecture |
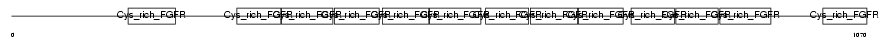
|
|||||
| Description | Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Selel). | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 3) | NC score | 0.050897 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
GAS8_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 4) | NC score | 0.087346 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 622 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q60779, Q99L71 | Gene names | Gas8, Gas11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
RABE1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 5) | NC score | 0.056862 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 6) | NC score | 0.048664 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 7) | NC score | 0.047913 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
GAK_HUMAN
|
||||||
| θ value | 0.163984 (rank : 8) | NC score | 0.024658 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
GAS8_HUMAN
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.064326 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
K1H5_HUMAN
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.031577 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92764, O76012, Q92651 | Gene names | KRTHA5, HHA5, HKA5 | |||
|
Domain Architecture |
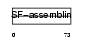
|
|||||
| Description | Keratin, type I cuticular Ha5 (Hair keratin, type I Ha5). | |||||
|
HAIR_MOUSE
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.038797 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61645, Q80Y47 | Gene names | Hr | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
K1H2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.027299 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14532 | Gene names | KRTHA2, HHA2, HKA2 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.042483 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
GAK_MOUSE
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.022522 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.041844 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
CSF1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.086765 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P07141, Q8R3C8 | Gene names | Csf1, Csfm | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF). | |||||
|
INAR2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.074649 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35664, O35238, O35663, O35983 | Gene names | Ifnar2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-alpha/beta receptor beta chain precursor (IFN-alpha-REC) (Type I interferon receptor) (IFN-R) (Interferon alpha/beta receptor 2). | |||||
|
CSF1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.069624 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
HDAC9_HUMAN
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.022360 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKV0, O94845, O95028 | Gene names | HDAC9, HDAC7, HDAC7B, KIAA0744 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 9 (HD9) (HD7B) (HD7). | |||||
|
IFIT3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.037986 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64345, Q99L62 | Gene names | Ifit3, Garg49, Ifi49 | |||
|
Domain Architecture |
|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 3 (IFIT-3) (Glucocorticoid-attenuated response gene 49 protein) (GARG-49) (IRG2). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.041679 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.041092 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.030005 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.042299 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
TDRD3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.030142 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H7E2, Q6P992 | Gene names | TDRD3 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 3. | |||||
|
HMMR_MOUSE
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.036592 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
CP250_MOUSE
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.034458 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
ETV4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.015109 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.030895 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
NUMB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.016667 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49757, Q6NUQ7, Q86SY1, Q8WW73, Q9UBG1, Q9UEQ4, Q9UKE8, Q9UKE9, Q9UKF0, Q9UQJ4 | Gene names | NUMB | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (h-Numb) (Protein S171). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.029473 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
QSCN6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.014445 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BND5, Q3TDY9, Q3TE19, Q3TR29, Q3UEL4, Q8K041, Q9DBL6 | Gene names | Qscn6, Sox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfhydryl oxidase 1 precursor (EC 1.8.3.2) (Quiescin Q6) (Skin sulfhydryl oxidase) (mSOx). | |||||
|
ZAN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.013209 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.026965 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CE152_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.035827 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
DC1L1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.020191 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R1Q8 | Gene names | Dync1li1, Dncli1, Dnclic1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 1 (Dynein light intermediate chain 1, cytosolic) (Dynein light chain A) (DLC-A). | |||||
|
DDX24_HUMAN
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.010860 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
TOR3A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.011919 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ER38, Q3T9Q6, Q3TDA9, Q3U5X1, Q3U8F2, Q9CR17, Q9CU67 | Gene names | Tor3a, Adir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-3A precursor (Torsin family 3 member A) (ATP-dependent interferon-responsive protein). | |||||
|
VAPB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.014834 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QY76 | Gene names | Vapb | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein B (VAMP- associated protein B) (VAMP-associated protein 33b) (VAMP-B) (VAP-B). | |||||
|
ABCA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.004581 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99758, Q54A95, Q92473 | Gene names | ABCA3, ABC3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette sub-family A member 3 (ATP-binding cassette transporter 3) (ATP-binding cassette 3) (ABC-C transporter). | |||||
|
EHMT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.004383 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
ETV4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.011909 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.027776 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
IFT81_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.029571 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
IL18R_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.012313 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61098 | Gene names | Il18r1 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-18 receptor 1 precursor (IL1 receptor-related protein) (IL-1Rrp). | |||||
|
NAL10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.011070 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86W26, Q6JGT0 | Gene names | NALP10, NOD8, PYNOD | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10 (Nucleotide-binding oligomerization domain protein 8). | |||||
|
RORG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.007617 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51449, Q8N5V7, Q8NCY8 | Gene names | RORC, NR1F3, RORG, RZRG | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor ROR-gamma (Nuclear receptor RZR-gamma). | |||||
|
SKIL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.033535 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60665, Q60702, Q78E90, Q80VK5 | Gene names | Skil, Skir, Sno | |||
|
Domain Architecture |

|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
T22D4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.011280 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.003140 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
GSLG1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61543, Q9QZ40 | Gene names | Glg1, Esl1, Mg160, Selel | |||
|
Domain Architecture |
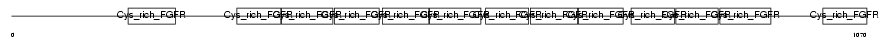
|
|||||
| Description | Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Selel). | |||||
|
GSLG1_HUMAN
|
||||||
| NC score | 0.995172 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q92896, Q13221 | Gene names | GLG1, CFR1, ESL1, MG160 | |||
|
Domain Architecture |
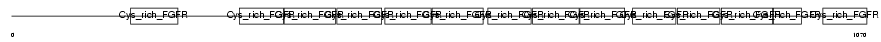
|
|||||
| Description | Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Cysteine-rich fibroblast growth factor receptor) (CFR-1). | |||||
|
GAS8_MOUSE
|
||||||
| NC score | 0.087346 (rank : 3) | θ value | 0.0330416 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 622 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q60779, Q99L71 | Gene names | Gas8, Gas11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
CSF1_MOUSE
|
||||||
| NC score | 0.086765 (rank : 4) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P07141, Q8R3C8 | Gene names | Csf1, Csfm | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF). | |||||
|
INAR2_MOUSE
|
||||||
| NC score | 0.074649 (rank : 5) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35664, O35238, O35663, O35983 | Gene names | Ifnar2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-alpha/beta receptor beta chain precursor (IFN-alpha-REC) (Type I interferon receptor) (IFN-R) (Interferon alpha/beta receptor 2). | |||||
|
CSF1_HUMAN
|
||||||
| NC score | 0.069624 (rank : 6) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
GAS8_HUMAN
|
||||||
| NC score | 0.064326 (rank : 7) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.056862 (rank : 8) | θ value | 0.0736092 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.050897 (rank : 9) | θ value | 0.0113563 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.048664 (rank : 10) | θ value | 0.0736092 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.047913 (rank : 11) | θ value | 0.0961366 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.042483 (rank : 12) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.042299 (rank : 13) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.041844 (rank : 14) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.041679 (rank : 15) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.041092 (rank : 16) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
HAIR_MOUSE
|
||||||
| NC score | 0.038797 (rank : 17) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61645, Q80Y47 | Gene names | Hr | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
IFIT3_MOUSE
|
||||||
| NC score | 0.037986 (rank : 18) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64345, Q99L62 | Gene names | Ifit3, Garg49, Ifi49 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 3 (IFIT-3) (Glucocorticoid-attenuated response gene 49 protein) (GARG-49) (IRG2). | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.036592 (rank : 19) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.035827 (rank : 20) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.034458 (rank : 21) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
SKIL_MOUSE
|
||||||
| NC score | 0.033535 (rank : 22) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60665, Q60702, Q78E90, Q80VK5 | Gene names | Skil, Skir, Sno | |||
|
Domain Architecture |

|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
K1H5_HUMAN
|
||||||
| NC score | 0.031577 (rank : 23) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92764, O76012, Q92651 | Gene names | KRTHA5, HHA5, HKA5 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha5 (Hair keratin, type I Ha5). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.030895 (rank : 24) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
TDRD3_HUMAN
|
||||||
| NC score | 0.030142 (rank : 25) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H7E2, Q6P992 | Gene names | TDRD3 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 3. | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.030005 (rank : 26) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
IFT81_HUMAN
|
||||||
| NC score | 0.029571 (rank : 27) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.029473 (rank : 28) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.027776 (rank : 29) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
K1H2_HUMAN
|
||||||
| NC score | 0.027299 (rank : 30) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14532 | Gene names | KRTHA2, HHA2, HKA2 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.026965 (rank : 31) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
GAK_HUMAN
|
||||||
| NC score | 0.024658 (rank : 32) | θ value | 0.163984 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
GAK_MOUSE
|
||||||
| NC score | 0.022522 (rank : 33) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
HDAC9_HUMAN
|
||||||
| NC score | 0.022360 (rank : 34) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKV0, O94845, O95028 | Gene names | HDAC9, HDAC7, HDAC7B, KIAA0744 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 9 (HD9) (HD7B) (HD7). | |||||
|
DC1L1_MOUSE
|
||||||
| NC score | 0.020191 (rank : 35) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R1Q8 | Gene names | Dync1li1, Dncli1, Dnclic1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 1 (Dynein light intermediate chain 1, cytosolic) (Dynein light chain A) (DLC-A). | |||||
|
NUMB_HUMAN
|
||||||
| NC score | 0.016667 (rank : 36) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49757, Q6NUQ7, Q86SY1, Q8WW73, Q9UBG1, Q9UEQ4, Q9UKE8, Q9UKE9, Q9UKF0, Q9UQJ4 | Gene names | NUMB | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (h-Numb) (Protein S171). | |||||
|
ETV4_HUMAN
|
||||||
| NC score | 0.015109 (rank : 37) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
VAPB_MOUSE
|
||||||
| NC score | 0.014834 (rank : 38) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QY76 | Gene names | Vapb | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein B (VAMP- associated protein B) (VAMP-associated protein 33b) (VAMP-B) (VAP-B). | |||||
|
QSCN6_MOUSE
|
||||||
| NC score | 0.014445 (rank : 39) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BND5, Q3TDY9, Q3TE19, Q3TR29, Q3UEL4, Q8K041, Q9DBL6 | Gene names | Qscn6, Sox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfhydryl oxidase 1 precursor (EC 1.8.3.2) (Quiescin Q6) (Skin sulfhydryl oxidase) (mSOx). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.013209 (rank : 40) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
IL18R_MOUSE
|
||||||
| NC score | 0.012313 (rank : 41) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61098 | Gene names | Il18r1 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-18 receptor 1 precursor (IL1 receptor-related protein) (IL-1Rrp). | |||||
|
TOR3A_MOUSE
|
||||||
| NC score | 0.011919 (rank : 42) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ER38, Q3T9Q6, Q3TDA9, Q3U5X1, Q3U8F2, Q9CR17, Q9CU67 | Gene names | Tor3a, Adir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-3A precursor (Torsin family 3 member A) (ATP-dependent interferon-responsive protein). | |||||
|
ETV4_MOUSE
|
||||||
| NC score | 0.011909 (rank : 43) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
T22D4_HUMAN
|
||||||
| NC score | 0.011280 (rank : 44) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
NAL10_HUMAN
|
||||||
| NC score | 0.011070 (rank : 45) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86W26, Q6JGT0 | Gene names | NALP10, NOD8, PYNOD | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10 (Nucleotide-binding oligomerization domain protein 8). | |||||
|
DDX24_HUMAN
|
||||||
| NC score | 0.010860 (rank : 46) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
RORG_HUMAN
|
||||||
| NC score | 0.007617 (rank : 47) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51449, Q8N5V7, Q8NCY8 | Gene names | RORC, NR1F3, RORG, RZRG | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor ROR-gamma (Nuclear receptor RZR-gamma). | |||||
|
ABCA3_HUMAN
|
||||||
| NC score | 0.004581 (rank : 48) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99758, Q54A95, Q92473 | Gene names | ABCA3, ABC3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette sub-family A member 3 (ATP-binding cassette transporter 3) (ATP-binding cassette 3) (ABC-C transporter). | |||||
|
EHMT1_HUMAN
|
||||||
| NC score | 0.004383 (rank : 49) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.003140 (rank : 50) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||