Please be patient as the page loads
|
CSF1_MOUSE
|
||||||
| SwissProt Accessions | P07141, Q8R3C8 | Gene names | Csf1, Csfm | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CSF1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.825071 (rank : 2) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
CSF1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P07141, Q8R3C8 | Gene names | Csf1, Csfm | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 3) | NC score | 0.103514 (rank : 3) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 4) | NC score | 0.059990 (rank : 35) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
GPR64_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 5) | NC score | 0.032934 (rank : 77) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IZP9, O00406, Q8IWT2, Q8IZE4, Q8IZE5, Q8IZE6, Q8IZE7, Q8IZP3, Q8IZP4 | Gene names | GPR64, HE6, TM7LN2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (He6 receptor). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 6) | NC score | 0.050500 (rank : 61) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 7) | NC score | 0.081666 (rank : 13) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 8) | NC score | 0.038740 (rank : 71) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
TF3C2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 9) | NC score | 0.061875 (rank : 31) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WUA4, Q16632, Q9BWI7 | Gene names | GTF3C2, KIAA0011 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 2 (Transcription factor IIIC-subunit beta) (TF3C-beta) (TFIIIC 110 kDa subunit) (TFIIIC110). | |||||
|
ASXL1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 10) | NC score | 0.058078 (rank : 39) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P59598 | Gene names | Asxl1, Kiaa0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.059825 (rank : 36) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.072374 (rank : 18) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
4ET_MOUSE
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.052106 (rank : 56) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9EST3, Q9CSS3 | Gene names | Eif4enif1, Clast4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
RFIP1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.078450 (rank : 14) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
ZN750_MOUSE
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.065655 (rank : 26) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
ZO1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.032039 (rank : 79) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
GSLG1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.087029 (rank : 9) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92896, Q13221 | Gene names | GLG1, CFR1, ESL1, MG160 | |||
|
Domain Architecture |
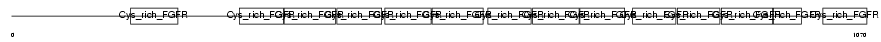
|
|||||
| Description | Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Cysteine-rich fibroblast growth factor receptor) (CFR-1). | |||||
|
GSLG1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.086765 (rank : 10) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61543, Q9QZ40 | Gene names | Glg1, Esl1, Mg160, Selel | |||
|
Domain Architecture |
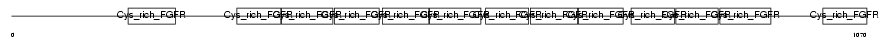
|
|||||
| Description | Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Selel). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.056734 (rank : 43) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.073770 (rank : 15) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.017607 (rank : 97) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
SYP2L_MOUSE
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.071696 (rank : 20) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
CK024_MOUSE
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.072786 (rank : 17) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.010401 (rank : 107) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.047871 (rank : 65) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
IL2RB_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.040108 (rank : 70) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14784 | Gene names | IL2RB | |||
|
Domain Architecture |
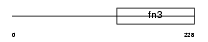
|
|||||
| Description | Interleukin-2 receptor beta chain precursor (IL-2 receptor) (P70-75) (p75) (High affinity IL-2 receptor subunit beta) (CD122 antigen). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.060216 (rank : 34) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NCA11_MOUSE
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.021875 (rank : 93) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
T240L_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.051818 (rank : 58) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q71F56, Q68DN4, Q9H8C0, Q9NSY9, Q9UFD8, Q9UPX5 | Gene names | THRAP2, KIAA1025, PROSIT240, TRAP240L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
T240L_MOUSE
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.053431 (rank : 52) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6JPI3, Q3TRF4, Q3UQI8, Q80TM3, Q80WQ0 | Gene names | Thrap2, Kiaa1025, Trap240l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.050568 (rank : 60) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.028659 (rank : 85) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.049828 (rank : 62) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.048405 (rank : 64) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
EPAS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.016649 (rank : 100) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
FBX42_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.029877 (rank : 83) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P3S6, Q5TEU8, Q86XI0, Q8N3N4, Q8N5F8, Q9BRM0, Q9P2L4 | Gene names | FBXO42, FBX42, KIAA1332 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 42. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.087970 (rank : 8) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.053982 (rank : 51) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.045704 (rank : 66) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
ZEP2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.013250 (rank : 104) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
CAC1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.013247 (rank : 105) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.081997 (rank : 12) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
JIP3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.016577 (rank : 101) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ESN9, Q99KU7, Q9EQD8, Q9ESN7, Q9ESN8, Q9ESP0, Q9JLH2, Q9JLH3, Q9R0U7 | Gene names | Mapk8ip3, Jip3, Jsap1, Syd2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3) (JNK/SAPK-associated protein 1) (JSAP1) (Sunday driver 2). | |||||
|
PO6F1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.018014 (rank : 95) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
POGZ_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.008727 (rank : 110) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BZH4, Q4VA94, Q80TZ8, Q8C0K1, Q8K294 | Gene names | Pogz, Kiaa0461 | |||
|
Domain Architecture |

|
|||||
| Description | Pogo transposable element with ZNF domain. | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.027200 (rank : 88) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.013223 (rank : 106) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.041851 (rank : 67) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.037933 (rank : 72) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
CDSN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.030927 (rank : 81) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TPC1 | Gene names | Cdsn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor. | |||||
|
IOD3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.022597 (rank : 90) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55073, Q8WVN5 | Gene names | DIO3, ITDI3, TXDI3 | |||
|
Domain Architecture |

|
|||||
| Description | Type III iodothyronine deiodinase (EC 1.97.1.11) (Type-III 5' deiodinase) (DIOIII) (Type 3 DI) (5DIII). | |||||
|
MAGD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.017383 (rank : 98) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UNF1, O76058, Q5BJF3, Q8NAL6, Q9H218, Q9P0U9, Q9UM52 | Gene names | MAGED2, BCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D2 (MAGE-D2 antigen) (MAGE-D) (Breast cancer-associated gene 1 protein) (BCG-1) (11B6) (Hepatocellular carcinoma-associated protein JCL-1). | |||||
|
YETS2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.032886 (rank : 78) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
CF142_MOUSE
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.030463 (rank : 82) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5FW52, Q9D6X9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf142 homolog. | |||||
|
CO002_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.060600 (rank : 32) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
CSPG5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.029782 (rank : 84) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95196, Q71M39, Q71M40 | Gene names | CSPG5, CALEB, NGC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
GTSE1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.041698 (rank : 68) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NYZ3, Q53GX5, Q5R3I6, Q6DHX4, Q9BRE0, Q9UGZ9, Q9Y557 | Gene names | GTSE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (B99 homolog). | |||||
|
MB3L2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.028107 (rank : 86) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NHZ7 | Gene names | MBD3L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 3-like 2 (MBD3-like 2). | |||||
|
TENX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.009604 (rank : 108) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
UT14A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.016427 (rank : 102) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q640M1, Q6A094, Q6EJB4, Q6P6L1, Q9CZK8 | Gene names | Utp14a, JsdX, Kiaa0266 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Juvenile spermatogonial depletion-like X-linked protein) (Jsd-like X-linked protein). | |||||
|
AF17_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.017885 (rank : 96) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.063599 (rank : 30) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CDA7L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.017299 (rank : 99) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q922M5 | Gene names | Cdca7l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle-associated 7-like protein (Transcription factor RAM2). | |||||
|
CENPT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.033188 (rank : 76) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3TJM4, Q3TPY6, Q3UPD2, Q8BTH0, Q8BTP2, Q8R5E9 | Gene names | Cenpt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein T (CENP-T). | |||||
|
CLMN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.009264 (rank : 109) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.014686 (rank : 103) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
HSF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.022397 (rank : 91) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
K0319_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.025537 (rank : 89) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5SZV5, Q80U39 | Gene names | Kiaa0319 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0319 precursor. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.027902 (rank : 87) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
P5I11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.033267 (rank : 75) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14683 | Gene names | TP53I11, PIG11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p53-inducible protein 11 (p53-induced protein 11). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.006030 (rank : 113) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
TYRO3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.002591 (rank : 115) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q06418, O14953, Q86VR3 | Gene names | TYRO3, BYK, DTK, RSE, SKY | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor TYRO3 precursor (EC 2.7.10.1) (Tyrosine-protein kinase RSE) (Tyrosine-protein kinase SKY) (Tyrosine- protein kinase DTK) (Protein-tyrosine kinase byk). | |||||
|
ASPP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.040143 (rank : 69) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.020737 (rank : 94) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.052117 (rank : 55) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
FBLN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.006891 (rank : 112) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
LSP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.022079 (rank : 92) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P19973, P97339, Q04950, Q62022, Q62023, Q62024, Q8CD28, Q99L65 | Gene names | Lsp1, Pp52, S37, Wp34 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte-specific protein 1 (Protein pp52) (52 kDa phosphoprotein) (Lymphocyte-specific antigen WP34) (S37 protein). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.037206 (rank : 73) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.031935 (rank : 80) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NDE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.007940 (rank : 111) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NXR1, Q49AQ2 | Gene names | NDE1, NUDE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE). | |||||
|
PGFRB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.003188 (rank : 114) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 962 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09619, Q8N5L4 | Gene names | PDGFRB | |||
|
Domain Architecture |

|
|||||
| Description | Beta platelet-derived growth factor receptor precursor (EC 2.7.10.1) (PDGF-R-beta) (CD140b antigen). | |||||
|
PO121_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.049758 (rank : 63) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
VGLL4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.037121 (rank : 74) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80V24 | Gene names | Vgll4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor vestigial-like protein 4 (Vgl-4). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.054195 (rank : 49) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CK024_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.054031 (rank : 50) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CS007_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.055660 (rank : 45) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.054643 (rank : 47) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.070125 (rank : 22) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
IL3RB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.056560 (rank : 44) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P32927 | Gene names | CSF2RB, IL3RB, IL5RB | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen) (CDw131). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.071425 (rank : 21) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.072022 (rank : 19) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.052613 (rank : 54) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.052099 (rank : 57) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.057167 (rank : 42) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.093974 (rank : 5) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.064870 (rank : 29) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.058837 (rank : 38) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.053036 (rank : 53) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.084702 (rank : 11) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.097862 (rank : 4) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.066517 (rank : 25) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.091358 (rank : 7) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.065040 (rank : 27) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.051047 (rank : 59) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
RERE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.058933 (rank : 37) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.057740 (rank : 40) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.069745 (rank : 24) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.064885 (rank : 28) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.069864 (rank : 23) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SYN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.054401 (rank : 48) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
TARA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.055311 (rank : 46) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.060443 (rank : 33) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.073476 (rank : 16) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.091748 (rank : 6) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.057588 (rank : 41) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
CSF1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P07141, Q8R3C8 | Gene names | Csf1, Csfm | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF). | |||||
|
CSF1_HUMAN
|
||||||
| NC score | 0.825071 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.103514 (rank : 3) | θ value | 0.00509761 (rank : 3) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.097862 (rank : 4) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.093974 (rank : 5) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.091748 (rank : 6) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.091358 (rank : 7) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.087970 (rank : 8) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
GSLG1_HUMAN
|
||||||
| NC score | 0.087029 (rank : 9) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92896, Q13221 | Gene names | GLG1, CFR1, ESL1, MG160 | |||
|
Domain Architecture |
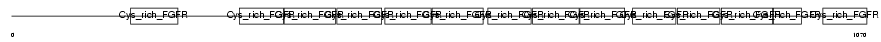
|
|||||
| Description | Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Cysteine-rich fibroblast growth factor receptor) (CFR-1). | |||||
|
GSLG1_MOUSE
|
||||||
| NC score | 0.086765 (rank : 10) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61543, Q9QZ40 | Gene names | Glg1, Esl1, Mg160, Selel | |||
|
Domain Architecture |
|
|||||
| Description | Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Selel). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.084702 (rank : 11) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.081997 (rank : 12) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.081666 (rank : 13) | θ value | 0.21417 (rank : 7) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
RFIP1_HUMAN
|
||||||
| NC score | 0.078450 (rank : 14) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.073770 (rank : 15) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.073476 (rank : 16) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
CK024_MOUSE
|
||||||
| NC score | 0.072786 (rank : 17) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.072374 (rank : 18) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.072022 (rank : 19) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
SYP2L_MOUSE
|
||||||
| NC score | 0.071696 (rank : 20) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.071425 (rank : 21) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.070125 (rank : 22) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.069864 (rank : 23) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.069745 (rank : 24) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.066517 (rank : 25) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
ZN750_MOUSE
|
||||||
| NC score | 0.065655 (rank : 26) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.065040 (rank : 27) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.064885 (rank : 28) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.064870 (rank : 29) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.063599 (rank : 30) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
TF3C2_HUMAN
|
||||||
| NC score | 0.061875 (rank : 31) | θ value | 0.279714 (rank : 9) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WUA4, Q16632, Q9BWI7 | Gene names | GTF3C2, KIAA0011 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 2 (Transcription factor IIIC-subunit beta) (TF3C-beta) (TFIIIC 110 kDa subunit) (TFIIIC110). | |||||
|
CO002_HUMAN
|
||||||
| NC score | 0.060600 (rank : 32) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.060443 (rank : 33) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.060216 (rank : 34) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.059990 (rank : 35) | θ value | 0.0431538 (rank : 4) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.059825 (rank : 36) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.058933 (rank : 37) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.058837 (rank : 38) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ASXL1_MOUSE
|
||||||
| NC score | 0.058078 (rank : 39) | θ value | 0.365318 (rank : 10) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P59598 | Gene names | Asxl1, Kiaa0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.057740 (rank : 40) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.057588 (rank : 41) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.057167 (rank : 42) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.056734 (rank : 43) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
IL3RB_HUMAN
|
||||||
| NC score | 0.056560 (rank : 44) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P32927 | Gene names | CSF2RB, IL3RB, IL5RB | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen) (CDw131). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.055660 (rank : 45) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.055311 (rank : 46) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.054643 (rank : 47) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.054401 (rank : 48) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.054195 (rank : 49) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.054031 (rank : 50) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.053982 (rank : 51) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
T240L_MOUSE
|
||||||
| NC score | 0.053431 (rank : 52) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6JPI3, Q3TRF4, Q3UQI8, Q80TM3, Q80WQ0 | Gene names | Thrap2, Kiaa1025, Trap240l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.053036 (rank : 53) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.052613 (rank : 54) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.052117 (rank : 55) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
4ET_MOUSE
|
||||||
| NC score | 0.052106 (rank : 56) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9EST3, Q9CSS3 | Gene names | Eif4enif1, Clast4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.052099 (rank : 57) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
T240L_HUMAN
|
||||||
| NC score | 0.051818 (rank : 58) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q71F56, Q68DN4, Q9H8C0, Q9NSY9, Q9UFD8, Q9UPX5 | Gene names | THRAP2, KIAA1025, PROSIT240, TRAP240L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.051047 (rank : 59) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.050568 (rank : 60) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.050500 (rank : 61) | θ value | 0.125558 (rank : 6) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.049828 (rank : 62) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.049758 (rank : 63) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.048405 (rank : 64) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.047871 (rank : 65) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.045704 (rank : 66) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.041851 (rank : 67) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
GTSE1_HUMAN
|
||||||
| NC score | 0.041698 (rank : 68) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NYZ3, Q53GX5, Q5R3I6, Q6DHX4, Q9BRE0, Q9UGZ9, Q9Y557 | Gene names | GTSE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (B99 homolog). | |||||
|
ASPP1_HUMAN
|
||||||
| NC score | 0.040143 (rank : 69) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
IL2RB_HUMAN
|
||||||
| NC score | 0.040108 (rank : 70) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14784 | Gene names | IL2RB | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-2 receptor beta chain precursor (IL-2 receptor) (P70-75) (p75) (High affinity IL-2 receptor subunit beta) (CD122 antigen). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.038740 (rank : 71) | θ value | 0.279714 (rank : 8) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.037933 (rank : 72) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.037206 (rank : 73) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
VGLL4_MOUSE
|
||||||
| NC score | 0.037121 (rank : 74) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80V24 | Gene names | Vgll4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor vestigial-like protein 4 (Vgl-4). | |||||
|
P5I11_HUMAN
|
||||||
| NC score | 0.033267 (rank : 75) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14683 | Gene names | TP53I11, PIG11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p53-inducible protein 11 (p53-induced protein 11). | |||||
|
CENPT_MOUSE
|
||||||
| NC score | 0.033188 (rank : 76) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3TJM4, Q3TPY6, Q3UPD2, Q8BTH0, Q8BTP2, Q8R5E9 | Gene names | Cenpt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein T (CENP-T). | |||||
|
GPR64_HUMAN
|
||||||
| NC score | 0.032934 (rank : 77) | θ value | 0.0736092 (rank : 5) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IZP9, O00406, Q8IWT2, Q8IZE4, Q8IZE5, Q8IZE6, Q8IZE7, Q8IZP3, Q8IZP4 | Gene names | GPR64, HE6, TM7LN2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (He6 receptor). | |||||
|
YETS2_HUMAN
|
||||||
| NC score | 0.032886 (rank : 78) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
ZO1_HUMAN
|
||||||
| NC score | 0.032039 (rank : 79) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.031935 (rank : 80) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
CDSN_MOUSE
|
||||||
| NC score | 0.030927 (rank : 81) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TPC1 | Gene names | Cdsn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor. | |||||
|
CF142_MOUSE
|
||||||
| NC score | 0.030463 (rank : 82) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5FW52, Q9D6X9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf142 homolog. | |||||
|
FBX42_HUMAN
|
||||||
| NC score | 0.029877 (rank : 83) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P3S6, Q5TEU8, Q86XI0, Q8N3N4, Q8N5F8, Q9BRM0, Q9P2L4 | Gene names | FBXO42, FBX42, KIAA1332 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 42. | |||||
|
CSPG5_HUMAN
|
||||||
| NC score | 0.029782 (rank : 84) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95196, Q71M39, Q71M40 | Gene names | CSPG5, CALEB, NGC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.028659 (rank : 85) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
MB3L2_HUMAN
|
||||||
| NC score | 0.028107 (rank : 86) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NHZ7 | Gene names | MBD3L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 3-like 2 (MBD3-like 2). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.027902 (rank : 87) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.027200 (rank : 88) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
K0319_MOUSE
|
||||||
| NC score | 0.025537 (rank : 89) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5SZV5, Q80U39 | Gene names | Kiaa0319 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0319 precursor. | |||||
|
IOD3_HUMAN
|
||||||
| NC score | 0.022597 (rank : 90) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55073, Q8WVN5 | Gene names | DIO3, ITDI3, TXDI3 | |||
|
Domain Architecture |

|
|||||
| Description | Type III iodothyronine deiodinase (EC 1.97.1.11) (Type-III 5' deiodinase) (DIOIII) (Type 3 DI) (5DIII). | |||||
|
HSF1_MOUSE
|
||||||
| NC score | 0.022397 (rank : 91) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
LSP1_MOUSE
|
||||||
| NC score | 0.022079 (rank : 92) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P19973, P97339, Q04950, Q62022, Q62023, Q62024, Q8CD28, Q99L65 | Gene names | Lsp1, Pp52, S37, Wp34 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte-specific protein 1 (Protein pp52) (52 kDa phosphoprotein) (Lymphocyte-specific antigen WP34) (S37 protein). | |||||
|
NCA11_MOUSE
|
||||||
| NC score | 0.021875 (rank : 93) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.020737 (rank : 94) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
PO6F1_MOUSE
|
||||||
| NC score | 0.018014 (rank : 95) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
AF17_HUMAN
|
||||||
| NC score | 0.017885 (rank : 96) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.017607 (rank : 97) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MAGD2_HUMAN
|
||||||
| NC score | 0.017383 (rank : 98) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UNF1, O76058, Q5BJF3, Q8NAL6, Q9H218, Q9P0U9, Q9UM52 | Gene names | MAGED2, BCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D2 (MAGE-D2 antigen) (MAGE-D) (Breast cancer-associated gene 1 protein) (BCG-1) (11B6) (Hepatocellular carcinoma-associated protein JCL-1). | |||||
|
CDA7L_MOUSE
|
||||||
| NC score | 0.017299 (rank : 99) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q922M5 | Gene names | Cdca7l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle-associated 7-like protein (Transcription factor RAM2). | |||||
|
EPAS1_MOUSE
|
||||||
| NC score | 0.016649 (rank : 100) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
JIP3_MOUSE
|
||||||
| NC score | 0.016577 (rank : 101) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ESN9, Q99KU7, Q9EQD8, Q9ESN7, Q9ESN8, Q9ESP0, Q9JLH2, Q9JLH3, Q9R0U7 | Gene names | Mapk8ip3, Jip3, Jsap1, Syd2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3) (JNK/SAPK-associated protein 1) (JSAP1) (Sunday driver 2). | |||||
|
UT14A_MOUSE
|
||||||
| NC score | 0.016427 (rank : 102) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q640M1, Q6A094, Q6EJB4, Q6P6L1, Q9CZK8 | Gene names | Utp14a, JsdX, Kiaa0266 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Juvenile spermatogonial depletion-like X-linked protein) (Jsd-like X-linked protein). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.014686 (rank : 103) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
ZEP2_MOUSE
|
||||||
| NC score | 0.013250 (rank : 104) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
CAC1A_HUMAN
|
||||||
| NC score | 0.013247 (rank : 105) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | 0.013223 (rank : 106) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
MAST2_HUMAN
|
||||||
| NC score | 0.010401 (rank : 107) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
TENX_HUMAN
|
||||||
| NC score | 0.009604 (rank : 108) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
CLMN_MOUSE
|
||||||
| NC score | 0.009264 (rank : 109) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
POGZ_MOUSE
|
||||||
| NC score | 0.008727 (rank : 110) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BZH4, Q4VA94, Q80TZ8, Q8C0K1, Q8K294 | Gene names | Pogz, Kiaa0461 | |||
|
Domain Architecture |

|
|||||
| Description | Pogo transposable element with ZNF domain. | |||||
|
NDE1_HUMAN
|
||||||
| NC score | 0.007940 (rank : 111) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NXR1, Q49AQ2 | Gene names | NDE1, NUDE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE). | |||||
|
FBLN2_MOUSE
|
||||||
| NC score | 0.006891 (rank : 112) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.006030 (rank : 113) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
PGFRB_HUMAN
|
||||||
| NC score | 0.003188 (rank : 114) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 962 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09619, Q8N5L4 | Gene names | PDGFRB | |||
|
Domain Architecture |

|
|||||
| Description | Beta platelet-derived growth factor receptor precursor (EC 2.7.10.1) (PDGF-R-beta) (CD140b antigen). | |||||
|
TYRO3_HUMAN
|
||||||
| NC score | 0.002591 (rank : 115) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q06418, O14953, Q86VR3 | Gene names | TYRO3, BYK, DTK, RSE, SKY | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor TYRO3 precursor (EC 2.7.10.1) (Tyrosine-protein kinase RSE) (Tyrosine-protein kinase SKY) (Tyrosine- protein kinase DTK) (Protein-tyrosine kinase byk). | |||||