Please be patient as the page loads
|
DDX20_MOUSE
|
||||||
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DDX20_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.995100 (rank : 2) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
DDX20_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 177 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DDX39_MOUSE
|
||||||
| θ value | 6.99855e-61 (rank : 3) | NC score | 0.966627 (rank : 3) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q8VDW0, Q3UJV4, Q8C2C2 | Gene names | Ddx39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39). | |||||
|
UAP56_HUMAN
|
||||||
| θ value | 1.31987e-59 (rank : 4) | NC score | 0.966198 (rank : 5) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q13838, O43496, Q5RJ64, Q5RJ66, Q5ST94, Q5STB4, Q5STB5, Q5STB7, Q5STU4, Q5STU5, Q5STU6, Q5STU8, Q71V76 | Gene names | BAT1, UAP56 | |||
|
Domain Architecture |

|
|||||
| Description | Spliceosome RNA helicase BAT1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (ATP-dependent RNA helicase p47) (HLA-B-associated transcript-1). | |||||
|
UAP56_MOUSE
|
||||||
| θ value | 1.31987e-59 (rank : 5) | NC score | 0.966169 (rank : 6) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9Z1N5, Q8HW97 | Gene names | Bat1, Bat1a, Uap56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spliceosome RNA helicase Bat1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (HLA-B-associated transcript 1). | |||||
|
DDX39_HUMAN
|
||||||
| θ value | 1.7238e-59 (rank : 6) | NC score | 0.966466 (rank : 4) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O00148, Q9BVP6, Q9H5W0 | Gene names | DDX39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39) (Nuclear RNA helicase URH49). | |||||
|
DDX6_HUMAN
|
||||||
| θ value | 4.24587e-58 (rank : 7) | NC score | 0.965409 (rank : 7) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
DDX6_MOUSE
|
||||||
| θ value | 4.24587e-58 (rank : 8) | NC score | 0.965187 (rank : 8) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
IF4A2_HUMAN
|
||||||
| θ value | 9.45883e-58 (rank : 9) | NC score | 0.963405 (rank : 9) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| θ value | 9.45883e-58 (rank : 10) | NC score | 0.963405 (rank : 10) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
DDX48_MOUSE
|
||||||
| θ value | 2.7521e-57 (rank : 11) | NC score | 0.962763 (rank : 14) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
DDX48_HUMAN
|
||||||
| θ value | 3.59435e-57 (rank : 12) | NC score | 0.962817 (rank : 13) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
IF4A1_HUMAN
|
||||||
| θ value | 4.69435e-57 (rank : 13) | NC score | 0.963082 (rank : 11) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| θ value | 4.69435e-57 (rank : 14) | NC score | 0.963082 (rank : 12) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
DD19B_HUMAN
|
||||||
| θ value | 8.02596e-49 (rank : 15) | NC score | 0.951669 (rank : 16) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
DD19A_HUMAN
|
||||||
| θ value | 3.04986e-48 (rank : 16) | NC score | 0.951941 (rank : 15) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19A_MOUSE
|
||||||
| θ value | 3.04986e-48 (rank : 17) | NC score | 0.951643 (rank : 17) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DDX25_HUMAN
|
||||||
| θ value | 2.95404e-43 (rank : 18) | NC score | 0.949327 (rank : 19) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX25_MOUSE
|
||||||
| θ value | 3.26607e-42 (rank : 19) | NC score | 0.948440 (rank : 20) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX47_MOUSE
|
||||||
| θ value | 4.71619e-41 (rank : 20) | NC score | 0.949368 (rank : 18) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX49_MOUSE
|
||||||
| θ value | 3.99248e-40 (rank : 21) | NC score | 0.945582 (rank : 22) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX47_HUMAN
|
||||||
| θ value | 6.81017e-40 (rank : 22) | NC score | 0.948270 (rank : 21) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX49_HUMAN
|
||||||
| θ value | 4.41421e-39 (rank : 23) | NC score | 0.940806 (rank : 23) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX27_MOUSE
|
||||||
| θ value | 6.37413e-38 (rank : 24) | NC score | 0.940252 (rank : 24) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX27_HUMAN
|
||||||
| θ value | 8.32485e-38 (rank : 25) | NC score | 0.938748 (rank : 25) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX43_HUMAN
|
||||||
| θ value | 3.16345e-37 (rank : 26) | NC score | 0.933127 (rank : 28) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
DDX10_MOUSE
|
||||||
| θ value | 7.79185e-36 (rank : 27) | NC score | 0.921980 (rank : 45) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX17_HUMAN
|
||||||
| θ value | 1.46948e-34 (rank : 28) | NC score | 0.924821 (rank : 43) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
DDX17_MOUSE
|
||||||
| θ value | 1.46948e-34 (rank : 29) | NC score | 0.924943 (rank : 42) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
DDX53_HUMAN
|
||||||
| θ value | 5.584e-34 (rank : 30) | NC score | 0.927967 (rank : 35) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
DDX54_MOUSE
|
||||||
| θ value | 7.29293e-34 (rank : 31) | NC score | 0.936340 (rank : 26) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
DDX23_HUMAN
|
||||||
| θ value | 2.12192e-33 (rank : 32) | NC score | 0.907490 (rank : 59) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
DDX41_MOUSE
|
||||||
| θ value | 2.77131e-33 (rank : 33) | NC score | 0.932241 (rank : 31) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
DDX41_HUMAN
|
||||||
| θ value | 4.72714e-33 (rank : 34) | NC score | 0.932768 (rank : 29) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
DDX3Y_HUMAN
|
||||||
| θ value | 1.0531e-32 (rank : 35) | NC score | 0.919413 (rank : 52) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
DDX3X_HUMAN
|
||||||
| θ value | 1.37539e-32 (rank : 36) | NC score | 0.919980 (rank : 48) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX3X_MOUSE
|
||||||
| θ value | 1.37539e-32 (rank : 37) | NC score | 0.919595 (rank : 51) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
PL10_MOUSE
|
||||||
| θ value | 1.79631e-32 (rank : 38) | NC score | 0.920275 (rank : 47) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
DDX54_HUMAN
|
||||||
| θ value | 2.34606e-32 (rank : 39) | NC score | 0.932194 (rank : 32) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
DDX10_HUMAN
|
||||||
| θ value | 4.00176e-32 (rank : 40) | NC score | 0.919301 (rank : 53) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX4_MOUSE
|
||||||
| θ value | 4.00176e-32 (rank : 41) | NC score | 0.927970 (rank : 34) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
DDX4_HUMAN
|
||||||
| θ value | 5.22648e-32 (rank : 42) | NC score | 0.926817 (rank : 37) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
DDX3Y_MOUSE
|
||||||
| θ value | 9.8567e-31 (rank : 43) | NC score | 0.918636 (rank : 54) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
DDX46_HUMAN
|
||||||
| θ value | 1.85889e-29 (rank : 44) | NC score | 0.908426 (rank : 57) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX46_MOUSE
|
||||||
| θ value | 1.85889e-29 (rank : 45) | NC score | 0.906627 (rank : 60) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
DDX52_HUMAN
|
||||||
| θ value | 1.85889e-29 (rank : 46) | NC score | 0.933760 (rank : 27) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX5_HUMAN
|
||||||
| θ value | 5.40856e-29 (rank : 47) | NC score | 0.919742 (rank : 49) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
DDX5_MOUSE
|
||||||
| θ value | 7.06379e-29 (rank : 48) | NC score | 0.919692 (rank : 50) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
DDX52_MOUSE
|
||||||
| θ value | 1.57365e-28 (rank : 49) | NC score | 0.932657 (rank : 30) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8K301, Q8BV29 | Gene names | Ddx52, Rok1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX55_HUMAN
|
||||||
| θ value | 1.73987e-27 (rank : 50) | NC score | 0.926134 (rank : 40) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 3.87602e-27 (rank : 51) | NC score | 0.905521 (rank : 61) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 8.63488e-27 (rank : 52) | NC score | 0.899193 (rank : 63) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX18_MOUSE
|
||||||
| θ value | 3.28125e-26 (rank : 53) | NC score | 0.924029 (rank : 44) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
DDX55_MOUSE
|
||||||
| θ value | 4.28545e-26 (rank : 54) | NC score | 0.926150 (rank : 39) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX18_HUMAN
|
||||||
| θ value | 2.35151e-24 (rank : 55) | NC score | 0.921108 (rank : 46) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
DDX50_MOUSE
|
||||||
| θ value | 2.35151e-24 (rank : 56) | NC score | 0.912392 (rank : 55) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX28_MOUSE
|
||||||
| θ value | 3.07116e-24 (rank : 57) | NC score | 0.929148 (rank : 33) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX50_HUMAN
|
||||||
| θ value | 5.23862e-24 (rank : 58) | NC score | 0.911599 (rank : 56) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX31_HUMAN
|
||||||
| θ value | 2.59989e-23 (rank : 59) | NC score | 0.908051 (rank : 58) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
DDX56_HUMAN
|
||||||
| θ value | 2.20094e-22 (rank : 60) | NC score | 0.926538 (rank : 38) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NY93, Q9H9I8 | Gene names | DDX56, DDX21, NOH61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase) (DEAD-box protein 21). | |||||
|
DDX28_HUMAN
|
||||||
| θ value | 6.40375e-22 (rank : 61) | NC score | 0.926972 (rank : 36) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX56_MOUSE
|
||||||
| θ value | 3.17815e-21 (rank : 62) | NC score | 0.925748 (rank : 41) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
DDX24_MOUSE
|
||||||
| θ value | 1.33526e-19 (rank : 63) | NC score | 0.900362 (rank : 62) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX24_HUMAN
|
||||||
| θ value | 3.88503e-19 (rank : 64) | NC score | 0.896185 (rank : 65) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX51_HUMAN
|
||||||
| θ value | 6.62687e-19 (rank : 65) | NC score | 0.881474 (rank : 66) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX1_HUMAN
|
||||||
| θ value | 1.92812e-18 (rank : 66) | NC score | 0.871908 (rank : 68) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
DDX1_MOUSE
|
||||||
| θ value | 2.5182e-18 (rank : 67) | NC score | 0.873147 (rank : 67) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
DDX51_MOUSE
|
||||||
| θ value | 2.5182e-18 (rank : 68) | NC score | 0.896624 (rank : 64) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
BLM_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 69) | NC score | 0.479093 (rank : 71) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O88700, O88198 | Gene names | Blm | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein homolog (EC 3.6.1.-) (mBLM). | |||||
|
BLM_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 70) | NC score | 0.463321 (rank : 72) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
NOL8_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 71) | NC score | 0.042204 (rank : 103) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
DICER_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 72) | NC score | 0.399831 (rank : 78) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
RECQ4_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 73) | NC score | 0.505195 (rank : 70) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
DICER_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 74) | NC score | 0.395905 (rank : 79) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
RECQ4_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 75) | NC score | 0.517453 (rank : 69) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
OXR1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 76) | NC score | 0.026666 (rank : 111) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
FANCM_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 77) | NC score | 0.410098 (rank : 75) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
NCLN_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 78) | NC score | 0.047569 (rank : 98) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCM8, Q8C7Y4, Q9CX81 | Gene names | Ncln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nicalin precursor (Nicastrin-like protein). | |||||
|
PHF1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 79) | NC score | 0.020489 (rank : 118) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
FANCM_MOUSE
|
||||||
| θ value | 0.125558 (rank : 80) | NC score | 0.403114 (rank : 77) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 81) | NC score | 0.055053 (rank : 93) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 82) | NC score | 0.054672 (rank : 94) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 83) | NC score | 0.043873 (rank : 101) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 84) | NC score | 0.044459 (rank : 100) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
NCLN_HUMAN
|
||||||
| θ value | 0.21417 (rank : 85) | NC score | 0.042936 (rank : 102) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q969V3, O75252, Q6FI60, Q6ZMB7, Q8TAT7, Q96H48, Q96IS7, Q9BQH9, Q9BTX4, Q9NPP2 | Gene names | NCLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nicalin precursor (Nicastrin-like protein). | |||||
|
PPHLN_HUMAN
|
||||||
| θ value | 0.21417 (rank : 86) | NC score | 0.029456 (rank : 107) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NEY8, Q86YT2, Q8IXN3, Q8TB09, Q96NB9, Q9NXL4, Q9P0P6, Q9P0R9 | Gene names | PPHLN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Periphilin-1 (Gastric cancer antigen Ga50). | |||||
|
CC026_MOUSE
|
||||||
| θ value | 0.279714 (rank : 87) | NC score | 0.166551 (rank : 88) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CZT6, Q9CY77 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26 homolog. | |||||
|
ENL_HUMAN
|
||||||
| θ value | 0.279714 (rank : 88) | NC score | 0.024548 (rank : 113) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
LMBL3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 89) | NC score | 0.012695 (rank : 135) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 90) | NC score | 0.008406 (rank : 156) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
WRN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 91) | NC score | 0.431911 (rank : 73) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
ARI4A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 92) | NC score | 0.029643 (rank : 106) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 0.365318 (rank : 93) | NC score | 0.024573 (rank : 112) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
ENKUR_HUMAN
|
||||||
| θ value | 0.47712 (rank : 94) | NC score | 0.020520 (rank : 117) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TC29 | Gene names | C10orf63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enkurin. | |||||
|
K1543_HUMAN
|
||||||
| θ value | 0.47712 (rank : 95) | NC score | 0.015504 (rank : 127) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 96) | NC score | 0.011003 (rank : 141) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 97) | NC score | 0.426346 (rank : 74) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 98) | NC score | 0.047690 (rank : 97) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
WRN_MOUSE
|
||||||
| θ value | 0.47712 (rank : 99) | NC score | 0.404485 (rank : 76) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 100) | NC score | 0.019448 (rank : 120) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
EXO1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 101) | NC score | 0.017088 (rank : 124) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UQ84, O60545, O75214, O75466, Q5T396, Q96IJ1, Q9UG38, Q9UNW0 | Gene names | EXO1, EXOI, HEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (hExo1) (Exonuclease I) (hExoI). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 102) | NC score | 0.032558 (rank : 105) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 0.62314 (rank : 103) | NC score | 0.021130 (rank : 116) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 104) | NC score | 0.018634 (rank : 121) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
SOCS6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 105) | NC score | 0.009631 (rank : 148) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14544 | Gene names | SOCS6, CIS4, SOCS4 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 6 (SOCS-6) (Suppressor of cytokine signaling 4) (SOCS-4) (Cytokine-inducible SH2 protein 4) (CIS-4). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 106) | NC score | 0.026780 (rank : 110) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
CC026_HUMAN
|
||||||
| θ value | 1.06291 (rank : 107) | NC score | 0.193972 (rank : 86) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BQ75 | Gene names | C3orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26. | |||||
|
CDN1C_MOUSE
|
||||||
| θ value | 1.06291 (rank : 108) | NC score | 0.016559 (rank : 125) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
PPHLN_MOUSE
|
||||||
| θ value | 1.06291 (rank : 109) | NC score | 0.028710 (rank : 108) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2H1, Q8BUZ6 | Gene names | Pphln1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Periphilin-1. | |||||
|
TSH1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 110) | NC score | 0.014007 (rank : 130) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
TTC14_HUMAN
|
||||||
| θ value | 1.06291 (rank : 111) | NC score | 0.013667 (rank : 132) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
AKAP2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 112) | NC score | 0.011226 (rank : 139) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 113) | NC score | 0.003047 (rank : 181) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
EXOC3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 114) | NC score | 0.013828 (rank : 131) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60645, Q8TEN6, Q8WUW0, Q96DI4 | Gene names | EXOC3, SEC6, SEC6L1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 3 (Exocyst complex component Sec6). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 1.38821 (rank : 115) | NC score | 0.018079 (rank : 122) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
IFIH1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 116) | NC score | 0.304202 (rank : 82) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 1.38821 (rank : 117) | NC score | 0.023510 (rank : 114) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
NPM_HUMAN
|
||||||
| θ value | 1.38821 (rank : 118) | NC score | 0.021799 (rank : 115) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P06748, P08693, Q12826, Q13440, Q13441, Q14115, Q5EU94, Q5EU95, Q5EU96, Q5EU97, Q5EU98, Q5EU99, Q6V962, Q8WTW5, Q96AT6, Q96DC4, Q96EA5 | Gene names | NPM1, NPM | |||
|
Domain Architecture |

|
|||||
| Description | Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38). | |||||
|
SYN3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 119) | NC score | 0.009175 (rank : 150) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8JZP2 | Gene names | Syn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-3 (Synapsin III). | |||||
|
BCL7A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 120) | NC score | 0.017405 (rank : 123) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q4VC05, Q13843 | Gene names | BCL7A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member A. | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 121) | NC score | 0.005817 (rank : 164) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
TTC28_HUMAN
|
||||||
| θ value | 1.81305 (rank : 122) | NC score | 0.008422 (rank : 155) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
UBQL3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 123) | NC score | 0.007861 (rank : 159) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C5U9 | Gene names | Ubqln3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 124) | NC score | 0.009388 (rank : 149) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 125) | NC score | 0.026903 (rank : 109) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
GAB1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 126) | NC score | 0.008087 (rank : 158) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 2.36792 (rank : 127) | NC score | 0.015289 (rank : 128) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 128) | NC score | 0.007543 (rank : 160) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
TAB3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 129) | NC score | 0.010330 (rank : 145) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
ABCA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 130) | NC score | 0.002344 (rank : 183) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BZC7 | Gene names | ABCA2, ABC2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette sub-family A member 2 (ATP-binding cassette transporter 2) (ATP-binding cassette 2). | |||||
|
BCL7A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 131) | NC score | 0.015669 (rank : 126) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CXE2, Q8C361, Q8C8M8, Q8VD15 | Gene names | Bcl7a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member A. | |||||
|
CO039_HUMAN
|
||||||
| θ value | 3.0926 (rank : 132) | NC score | 0.008808 (rank : 153) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
MECP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 133) | NC score | 0.009985 (rank : 146) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51608, O15233 | Gene names | MECP2 | |||
|
Domain Architecture |
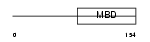
|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 134) | NC score | 0.020468 (rank : 119) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 135) | NC score | 0.004610 (rank : 170) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
APC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 136) | NC score | 0.011919 (rank : 137) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
APC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 137) | NC score | 0.012275 (rank : 136) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
BBX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 138) | NC score | 0.008482 (rank : 154) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WY36, Q2TAJ1, Q7L3J8, Q7LBY8, Q8NDB0, Q8WY35, Q9H0J6 | Gene names | BBX, HBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
CDT1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 139) | NC score | 0.009092 (rank : 151) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H211, Q86XX9, Q96CJ5, Q96GK5, Q96H67, Q96HE6, Q9BWM0 | Gene names | CDT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication factor Cdt1 (Double parked homolog) (DUP). | |||||
|
CYTSA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 140) | NC score | 0.000799 (rank : 185) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
JHD2C_MOUSE
|
||||||
| θ value | 4.03905 (rank : 141) | NC score | 0.011328 (rank : 138) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 142) | NC score | 0.005270 (rank : 167) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
NPDC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 143) | NC score | 0.010992 (rank : 142) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQX5, Q9BTD6, Q9BXT3, Q9NQS2, Q9Y434 | Gene names | NPDC1 | |||
|
Domain Architecture |
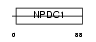
|
|||||
| Description | Neural proliferation differentiation and control protein 1 precursor (NPDC-1 protein). | |||||
|
TIRAP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 144) | NC score | 0.009839 (rank : 147) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99JY1, Q91ZW0 | Gene names | Tirap, Mal | |||
|
Domain Architecture |
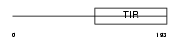
|
|||||
| Description | Toll-interleukin 1 receptor domain-containing adapter protein (TIR domain-containing adapter protein) (MyD88 adapter-like protein) (Adaptor protein Wyatt). | |||||
|
BBX_MOUSE
|
||||||
| θ value | 5.27518 (rank : 145) | NC score | 0.006982 (rank : 161) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VBW5, Q3TZK1, Q6NXY8, Q6PEU3, Q8BQJ7, Q8C7E0, Q8CDQ0, Q8CDV1, Q8VI48, Q8VI49, Q8VI50, Q9CS94 | Gene names | Bbx, Hbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 146) | NC score | 0.004890 (rank : 168) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 147) | NC score | 0.045496 (rank : 99) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 5.27518 (rank : 148) | NC score | 0.014119 (rank : 129) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
IFIH1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 149) | NC score | 0.285202 (rank : 84) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
MITF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 150) | NC score | 0.004207 (rank : 174) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 151) | NC score | 0.010592 (rank : 144) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PELI2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 152) | NC score | 0.004338 (rank : 172) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HAT8 | Gene names | PELI2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein pellino homolog 2 (Pellino-2). | |||||
|
PHF2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 153) | NC score | 0.004409 (rank : 171) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
RECQ1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 154) | NC score | 0.389293 (rank : 80) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P46063 | Gene names | RECQL, RECQL1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 5.27518 (rank : 155) | NC score | -0.005796 (rank : 186) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 156) | NC score | 0.011089 (rank : 140) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TMCC3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 157) | NC score | 0.003973 (rank : 175) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R310 | Gene names | Tmcc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 3. | |||||
|
EMSY_MOUSE
|
||||||
| θ value | 6.88961 (rank : 158) | NC score | 0.010843 (rank : 143) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
EP300_HUMAN
|
||||||
| θ value | 6.88961 (rank : 159) | NC score | 0.003271 (rank : 179) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
JERKY_HUMAN
|
||||||
| θ value | 6.88961 (rank : 160) | NC score | 0.005603 (rank : 165) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75564, O75565 | Gene names | JRK, JH8 | |||
|
Domain Architecture |
|
|||||
| Description | Jerky protein homolog. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 6.88961 (rank : 161) | NC score | 0.008859 (rank : 152) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RBMX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 162) | NC score | 0.003288 (rank : 178) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y388, Q5JY82, Q9Y3I8 | Gene names | RBMX2 | |||
|
Domain Architecture |
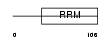
|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
RECQ1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 163) | NC score | 0.387292 (rank : 81) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 164) | NC score | 0.013447 (rank : 133) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TAB3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 165) | NC score | 0.008347 (rank : 157) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
ZCH10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 166) | NC score | 0.013053 (rank : 134) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
CHD5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.040135 (rank : 104) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CMGA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.006561 (rank : 163) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
DNL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.006582 (rank : 162) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
EYA4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.003050 (rank : 180) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z191 | Gene names | Eya4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
K2027_HUMAN
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.005503 (rank : 166) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
NFH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.004787 (rank : 169) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RBM28_MOUSE
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.002714 (rank : 182) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CGC6, Q8BG25, Q8VEJ8, Q9CS22, Q9CSE6 | Gene names | Rbm28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 28 (RNA-binding motif protein 28). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.001599 (rank : 184) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
TRA2A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | 0.003450 (rank : 176) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13595 | Gene names | TRA2A | |||
|
Domain Architecture |
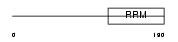
|
|||||
| Description | Transformer-2 protein homolog (TRA-2 alpha). | |||||
|
TRA2A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 176) | NC score | 0.003428 (rank : 177) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PFR5 | Gene names | Tra2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transformer-2 protein homolog (TRA-2 alpha). | |||||
|
UBP8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 177) | NC score | 0.004313 (rank : 173) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
DDX58_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.144430 (rank : 89) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
DDX58_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.173399 (rank : 87) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
HELC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.084360 (rank : 91) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
LGP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.282574 (rank : 85) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
LGP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.299150 (rank : 83) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
SMAL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.075706 (rank : 92) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
SMAL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.054122 (rank : 95) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BJL0, Q8BVK7, Q8BXW4, Q8K309, Q9EQK8, Q9QYC4 | Gene names | Smarcal1, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (mharp). | |||||
|
SMCA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.053908 (rank : 96) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PGB8, Q8BSS1, Q91Y58 | Gene names | Smarca1, Snf2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) (DNA-dependent ATPase SNF2L). | |||||
|
U520_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.108533 (rank : 90) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75643, O94884, Q6PX59, Q96IF2, Q9H7S0 | Gene names | ASCC3L1, HELIC2, KIAA0788 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U5 small nuclear ribonucleoprotein 200 kDa helicase (EC 3.6.1.-) (U5 snRNP-specific 200 kDa protein) (U5-200KD) (Activating signal cointegrator 1 complex subunit 3-like 1) (BRR2 homolog). | |||||
|
DDX20_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 177 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DDX20_HUMAN
|
||||||
| NC score | 0.995100 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
DDX39_MOUSE
|
||||||
| NC score | 0.966627 (rank : 3) | θ value | 6.99855e-61 (rank : 3) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q8VDW0, Q3UJV4, Q8C2C2 | Gene names | Ddx39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39). | |||||
|
DDX39_HUMAN
|
||||||
| NC score | 0.966466 (rank : 4) | θ value | 1.7238e-59 (rank : 6) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O00148, Q9BVP6, Q9H5W0 | Gene names | DDX39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39) (Nuclear RNA helicase URH49). | |||||
|
UAP56_HUMAN
|
||||||
| NC score | 0.966198 (rank : 5) | θ value | 1.31987e-59 (rank : 4) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q13838, O43496, Q5RJ64, Q5RJ66, Q5ST94, Q5STB4, Q5STB5, Q5STB7, Q5STU4, Q5STU5, Q5STU6, Q5STU8, Q71V76 | Gene names | BAT1, UAP56 | |||
|
Domain Architecture |

|
|||||
| Description | Spliceosome RNA helicase BAT1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (ATP-dependent RNA helicase p47) (HLA-B-associated transcript-1). | |||||
|
UAP56_MOUSE
|
||||||
| NC score | 0.966169 (rank : 6) | θ value | 1.31987e-59 (rank : 5) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9Z1N5, Q8HW97 | Gene names | Bat1, Bat1a, Uap56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spliceosome RNA helicase Bat1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (HLA-B-associated transcript 1). | |||||
|
DDX6_HUMAN
|
||||||
| NC score | 0.965409 (rank : 7) | θ value | 4.24587e-58 (rank : 7) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
DDX6_MOUSE
|
||||||
| NC score | 0.965187 (rank : 8) | θ value | 4.24587e-58 (rank : 8) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
IF4A2_HUMAN
|
||||||
| NC score | 0.963405 (rank : 9) | θ value | 9.45883e-58 (rank : 9) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| NC score | 0.963405 (rank : 10) | θ value | 9.45883e-58 (rank : 10) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A1_HUMAN
|
||||||
| NC score | 0.963082 (rank : 11) | θ value | 4.69435e-57 (rank : 13) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| NC score | 0.963082 (rank : 12) | θ value | 4.69435e-57 (rank : 14) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
DDX48_HUMAN
|
||||||
| NC score | 0.962817 (rank : 13) | θ value | 3.59435e-57 (rank : 12) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX48_MOUSE
|
||||||
| NC score | 0.962763 (rank : 14) | θ value | 2.7521e-57 (rank : 11) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
DD19A_HUMAN
|
||||||
| NC score | 0.951941 (rank : 15) | θ value | 3.04986e-48 (rank : 16) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19B_HUMAN
|
||||||
| NC score | 0.951669 (rank : 16) | θ value | 8.02596e-49 (rank : 15) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
DD19A_MOUSE
|
||||||
| NC score | 0.951643 (rank : 17) | θ value | 3.04986e-48 (rank : 17) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DDX47_MOUSE
|
||||||
| NC score | 0.949368 (rank : 18) | θ value | 4.71619e-41 (rank : 20) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX25_HUMAN
|
||||||
| NC score | 0.949327 (rank : 19) | θ value | 2.95404e-43 (rank : 18) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX25_MOUSE
|
||||||
| NC score | 0.948440 (rank : 20) | θ value | 3.26607e-42 (rank : 19) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX47_HUMAN
|
||||||
| NC score | 0.948270 (rank : 21) | θ value | 6.81017e-40 (rank : 22) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX49_MOUSE
|
||||||
| NC score | 0.945582 (rank : 22) | θ value | 3.99248e-40 (rank : 21) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX49_HUMAN
|
||||||
| NC score | 0.940806 (rank : 23) | θ value | 4.41421e-39 (rank : 23) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX27_MOUSE
|
||||||
| NC score | 0.940252 (rank : 24) | θ value | 6.37413e-38 (rank : 24) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX27_HUMAN
|
||||||
| NC score | 0.938748 (rank : 25) | θ value | 8.32485e-38 (rank : 25) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX54_MOUSE
|
||||||
| NC score | 0.936340 (rank : 26) | θ value | 7.29293e-34 (rank : 31) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
DDX52_HUMAN
|
||||||
| NC score | 0.933760 (rank : 27) | θ value | 1.85889e-29 (rank : 46) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX43_HUMAN
|
||||||
| NC score | 0.933127 (rank : 28) | θ value | 3.16345e-37 (rank : 26) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
DDX41_HUMAN
|
||||||
| NC score | 0.932768 (rank : 29) | θ value | 4.72714e-33 (rank : 34) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
DDX52_MOUSE
|
||||||
| NC score | 0.932657 (rank : 30) | θ value | 1.57365e-28 (rank : 49) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8K301, Q8BV29 | Gene names | Ddx52, Rok1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX41_MOUSE
|
||||||
| NC score | 0.932241 (rank : 31) | θ value | 2.77131e-33 (rank : 33) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
DDX54_HUMAN
|
||||||
| NC score | 0.932194 (rank : 32) | θ value | 2.34606e-32 (rank : 39) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
DDX28_MOUSE
|
||||||
| NC score | 0.929148 (rank : 33) | θ value | 3.07116e-24 (rank : 57) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX4_MOUSE
|
||||||
| NC score | 0.927970 (rank : 34) | θ value | 4.00176e-32 (rank : 41) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
DDX53_HUMAN
|
||||||
| NC score | 0.927967 (rank : 35) | θ value | 5.584e-34 (rank : 30) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
DDX28_HUMAN
|
||||||
| NC score | 0.926972 (rank : 36) | θ value | 6.40375e-22 (rank : 61) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX4_HUMAN
|
||||||
| NC score | 0.926817 (rank : 37) | θ value | 5.22648e-32 (rank : 42) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
DDX56_HUMAN
|
||||||
| NC score | 0.926538 (rank : 38) | θ value | 2.20094e-22 (rank : 60) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NY93, Q9H9I8 | Gene names | DDX56, DDX21, NOH61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase) (DEAD-box protein 21). | |||||
|
DDX55_MOUSE
|
||||||
| NC score | 0.926150 (rank : 39) | θ value | 4.28545e-26 (rank : 54) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX55_HUMAN
|
||||||
| NC score | 0.926134 (rank : 40) | θ value | 1.73987e-27 (rank : 50) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX56_MOUSE
|
||||||
| NC score | 0.925748 (rank : 41) | θ value | 3.17815e-21 (rank : 62) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
DDX17_MOUSE
|
||||||
| NC score | 0.924943 (rank : 42) | θ value | 1.46948e-34 (rank : 29) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
DDX17_HUMAN
|
||||||
| NC score | 0.924821 (rank : 43) | θ value | 1.46948e-34 (rank : 28) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
DDX18_MOUSE
|
||||||
| NC score | 0.924029 (rank : 44) | θ value | 3.28125e-26 (rank : 53) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
DDX10_MOUSE
|
||||||
| NC score | 0.921980 (rank : 45) | θ value | 7.79185e-36 (rank : 27) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX18_HUMAN
|
||||||
| NC score | 0.921108 (rank : 46) | θ value | 2.35151e-24 (rank : 55) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
PL10_MOUSE
|
||||||
| NC score | 0.920275 (rank : 47) | θ value | 1.79631e-32 (rank : 38) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
DDX3X_HUMAN
|
||||||
| NC score | 0.919980 (rank : 48) | θ value | 1.37539e-32 (rank : 36) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX5_HUMAN
|
||||||
| NC score | 0.919742 (rank : 49) | θ value | 5.40856e-29 (rank : 47) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
DDX5_MOUSE
|
||||||
| NC score | 0.919692 (rank : 50) | θ value | 7.06379e-29 (rank : 48) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
DDX3X_MOUSE
|
||||||
| NC score | 0.919595 (rank : 51) | θ value | 1.37539e-32 (rank : 37) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
DDX3Y_HUMAN
|
||||||
| NC score | 0.919413 (rank : 52) | θ value | 1.0531e-32 (rank : 35) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
DDX10_HUMAN
|
||||||
| NC score | 0.919301 (rank : 53) | θ value | 4.00176e-32 (rank : 40) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX3Y_MOUSE
|
||||||
| NC score | 0.918636 (rank : 54) | θ value | 9.8567e-31 (rank : 43) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
DDX50_MOUSE
|
||||||
| NC score | 0.912392 (rank : 55) | θ value | 2.35151e-24 (rank : 56) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX50_HUMAN
|
||||||
| NC score | 0.911599 (rank : 56) | θ value | 5.23862e-24 (rank : 58) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX46_HUMAN
|
||||||
| NC score | 0.908426 (rank : 57) | θ value | 1.85889e-29 (rank : 44) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX31_HUMAN
|
||||||
| NC score | 0.908051 (rank : 58) | θ value | 2.59989e-23 (rank : 59) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
DDX23_HUMAN
|
||||||
| NC score | 0.907490 (rank : 59) | θ value | 2.12192e-33 (rank : 32) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
DDX46_MOUSE
|
||||||
| NC score | 0.906627 (rank : 60) | θ value | 1.85889e-29 (rank : 45) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.905521 (rank : 61) | θ value | 3.87602e-27 (rank : 51) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX24_MOUSE
|
||||||
| NC score | 0.900362 (rank : 62) | θ value | 1.33526e-19 (rank : 63) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.899193 (rank : 63) | θ value | 8.63488e-27 (rank : 52) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX51_MOUSE
|
||||||
| NC score | 0.896624 (rank : 64) | θ value | 2.5182e-18 (rank : 68) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX24_HUMAN
|
||||||
| NC score | 0.896185 (rank : 65) | θ value | 3.88503e-19 (rank : 64) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX51_HUMAN
|
||||||
| NC score | 0.881474 (rank : 66) | θ value | 6.62687e-19 (rank : 65) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX1_MOUSE
|
||||||
| NC score | 0.873147 (rank : 67) | θ value | 2.5182e-18 (rank : 67) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
DDX1_HUMAN
|
||||||
| NC score | 0.871908 (rank : 68) | θ value | 1.92812e-18 (rank : 66) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
RECQ4_HUMAN
|
||||||
| NC score | 0.517453 (rank : 69) | θ value | 0.00665767 (rank : 75) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
RECQ4_MOUSE
|
||||||
| NC score | 0.505195 (rank : 70) | θ value | 0.00134147 (rank : 73) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
BLM_MOUSE
|
||||||
| NC score | 0.479093 (rank : 71) | θ value | 1.43324e-05 (rank : 69) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O88700, O88198 | Gene names | Blm | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein homolog (EC 3.6.1.-) (mBLM). | |||||
|
BLM_HUMAN
|
||||||
| NC score | 0.463321 (rank : 72) | θ value | 9.29e-05 (rank : 70) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
WRN_HUMAN
|
||||||
| NC score | 0.431911 (rank : 73) | θ value | 0.279714 (rank : 91) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.426346 (rank : 74) | θ value | 0.47712 (rank : 97) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
FANCM_HUMAN
|
||||||
| NC score | 0.410098 (rank : 75) | θ value | 0.0330416 (rank : 77) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
WRN_MOUSE
|
||||||
| NC score | 0.404485 (rank : 76) | θ value | 0.47712 (rank : 99) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
FANCM_MOUSE
|
||||||
| NC score | 0.403114 (rank : 77) | θ value | 0.125558 (rank : 80) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
DICER_HUMAN
|
||||||
| NC score | 0.399831 (rank : 78) | θ value | 0.00102713 (rank : 72) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
DICER_MOUSE
|
||||||
| NC score | 0.395905 (rank : 79) | θ value | 0.00298849 (rank : 74) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
RECQ1_HUMAN
|
||||||
| NC score | 0.389293 (rank : 80) | θ value | 5.27518 (rank : 154) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P46063 | Gene names | RECQL, RECQL1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
RECQ1_MOUSE
|
||||||
| NC score | 0.387292 (rank : 81) | θ value | 6.88961 (rank : 163) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
IFIH1_HUMAN
|
||||||
| NC score | 0.304202 (rank : 82) | θ value | 1.38821 (rank : 116) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
LGP2_MOUSE
|
||||||
| NC score | 0.299150 (rank : 83) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
IFIH1_MOUSE
|
||||||
| NC score | 0.285202 (rank : 84) | θ value | 5.27518 (rank : 149) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
LGP2_HUMAN
|
||||||
| NC score | 0.282574 (rank : 85) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
CC026_HUMAN
|
||||||
| NC score | 0.193972 (rank : 86) | θ value | 1.06291 (rank : 107) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BQ75 | Gene names | C3orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26. | |||||
|
DDX58_MOUSE
|
||||||
| NC score | 0.173399 (rank : 87) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
CC026_MOUSE
|
||||||
| NC score | 0.166551 (rank : 88) | θ value | 0.279714 (rank : 87) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CZT6, Q9CY77 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26 homolog. | |||||
|
DDX58_HUMAN
|
||||||
| NC score | 0.144430 (rank : 89) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
U520_HUMAN
|
||||||
| NC score | 0.108533 (rank : 90) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75643, O94884, Q6PX59, Q96IF2, Q9H7S0 | Gene names | ASCC3L1, HELIC2, KIAA0788 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U5 small nuclear ribonucleoprotein 200 kDa helicase (EC 3.6.1.-) (U5 snRNP-specific 200 kDa protein) (U5-200KD) (Activating signal cointegrator 1 complex subunit 3-like 1) (BRR2 homolog). | |||||
|
HELC1_HUMAN
|
||||||
| NC score | 0.084360 (rank : 91) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
SMAL1_HUMAN
|
||||||
| NC score | 0.075706 (rank : 92) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.055053 (rank : 93) | θ value | 0.163984 (rank : 81) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.054672 (rank : 94) | θ value | 0.163984 (rank : 82) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
SMAL1_MOUSE
|
||||||
| NC score | 0.054122 (rank : 95) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BJL0, Q8BVK7, Q8BXW4, Q8K309, Q9EQK8, Q9QYC4 | Gene names | Smarcal1, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (mharp). | |||||
|
SMCA1_MOUSE
|
||||||
| NC score | 0.053908 (rank : 96) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PGB8, Q8BSS1, Q91Y58 | Gene names | Smarca1, Snf2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) (DNA-dependent ATPase SNF2L). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.047690 (rank : 97) | θ value | 0.47712 (rank : 98) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
NCLN_MOUSE
|
||||||
| NC score | 0.047569 (rank : 98) | θ value | 0.0563607 (rank : 78) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCM8, Q8C7Y4, Q9CX81 | Gene names | Ncln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nicalin precursor (Nicastrin-like protein). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.045496 (rank : 99) | θ value | 5.27518 (rank : 147) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.044459 (rank : 100) | θ value | 0.21417 (rank : 84) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.043873 (rank : 101) | θ value | 0.21417 (rank : 83) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
NCLN_HUMAN
|
||||||
| NC score | 0.042936 (rank : 102) | θ value | 0.21417 (rank : 85) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q969V3, O75252, Q6FI60, Q6ZMB7, Q8TAT7, Q96H48, Q96IS7, Q9BQH9, Q9BTX4, Q9NPP2 | Gene names | NCLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nicalin precursor (Nicastrin-like protein). | |||||
|
NOL8_HUMAN
|
||||||
| NC score | 0.042204 (rank : 103) | θ value | 0.000461057 (rank : 71) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.040135 (rank : 104) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.032558 (rank : 105) | θ value | 0.62314 (rank : 102) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ARI4A_HUMAN
|
||||||
| NC score | 0.029643 (rank : 106) | θ value | 0.365318 (rank : 92) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
PPHLN_HUMAN
|
||||||
| NC score | 0.029456 (rank : 107) | θ value | 0.21417 (rank : 86) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NEY8, Q86YT2, Q8IXN3, Q8TB09, Q96NB9, Q9NXL4, Q9P0P6, Q9P0R9 | Gene names | PPHLN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Periphilin-1 (Gastric cancer antigen Ga50). | |||||
|
PPHLN_MOUSE
|
||||||
| NC score | 0.028710 (rank : 108) | θ value | 1.06291 (rank : 109) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2H1, Q8BUZ6 | Gene names | Pphln1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Periphilin-1. | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.026903 (rank : 109) | θ value | 2.36792 (rank : 125) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.026780 (rank : 110) | θ value | 0.813845 (rank : 106) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
OXR1_MOUSE
|
||||||
| NC score | 0.026666 (rank : 111) | θ value | 0.0252991 (rank : 76) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.024573 (rank : 112) | θ value | 0.365318 (rank : 93) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.024548 (rank : 113) | θ value | 0.279714 (rank : 88) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.023510 (rank : 114) | θ value | 1.38821 (rank : 117) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
NPM_HUMAN
|
||||||
| NC score | 0.021799 (rank : 115) | θ value | 1.38821 (rank : 118) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P06748, P08693, Q12826, Q13440, Q13441, Q14115, Q5EU94, Q5EU95, Q5EU96, Q5EU97, Q5EU98, Q5EU99, Q6V962, Q8WTW5, Q96AT6, Q96DC4, Q96EA5 | Gene names | NPM1, NPM | |||
|
Domain Architecture |

|
|||||
| Description | Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.021130 (rank : 116) | θ value | 0.62314 (rank : 103) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
ENKUR_HUMAN
|
||||||
| NC score | 0.020520 (rank : 117) | θ value | 0.47712 (rank : 94) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TC29 | Gene names | C10orf63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enkurin. | |||||
|
PHF1_HUMAN
|
||||||
| NC score | 0.020489 (rank : 118) | θ value | 0.0563607 (rank : 79) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.020468 (rank : 119) | θ value | 3.0926 (rank : 134) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.019448 (rank : 120) | θ value | 0.62314 (rank : 100) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.018634 (rank : 121) | θ value | 0.813845 (rank : 104) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.018079 (rank : 122) | θ value | 1.38821 (rank : 115) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
BCL7A_HUMAN
|
||||||
| NC score | 0.017405 (rank : 123) | θ value | 1.81305 (rank : 120) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q4VC05, Q13843 | Gene names | BCL7A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member A. | |||||
|
EXO1_HUMAN
|
||||||
| NC score | 0.017088 (rank : 124) | θ value | 0.62314 (rank : 101) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UQ84, O60545, O75214, O75466, Q5T396, Q96IJ1, Q9UG38, Q9UNW0 | Gene names | EXO1, EXOI, HEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (hExo1) (Exonuclease I) (hExoI). | |||||
|
CDN1C_MOUSE
|
||||||
| NC score | 0.016559 (rank : 125) | θ value | 1.06291 (rank : 108) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
BCL7A_MOUSE
|
||||||
| NC score | 0.015669 (rank : 126) | θ value | 3.0926 (rank : 131) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CXE2, Q8C361, Q8C8M8, Q8VD15 | Gene names | Bcl7a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member A. | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.015504 (rank : 127) | θ value | 0.47712 (rank : 95) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.015289 (rank : 128) | θ value | 2.36792 (rank : 127) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.014119 (rank : 129) | θ value | 5.27518 (rank : 148) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
TSH1_MOUSE
|
||||||
| NC score | 0.014007 (rank : 130) | θ value | 1.06291 (rank : 110) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
EXOC3_HUMAN
|
||||||
| NC score | 0.013828 (rank : 131) | θ value | 1.38821 (rank : 114) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60645, Q8TEN6, Q8WUW0, Q96DI4 | Gene names | EXOC3, SEC6, SEC6L1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 3 (Exocyst complex component Sec6). | |||||
|
TTC14_HUMAN
|
||||||
| NC score | 0.013667 (rank : 132) | θ value | 1.06291 (rank : 111) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.013447 (rank : 133) | θ value | 6.88961 (rank : 164) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ZCH10_MOUSE
|
||||||
| NC score | 0.013053 (rank : 134) | θ value | 6.88961 (rank : 166) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
LMBL3_MOUSE
|
||||||
| NC score | 0.012695 (rank : 135) | θ value | 0.279714 (rank : 89) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.012275 (rank : 136) | θ value | 4.03905 (rank : 137) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.011919 (rank : 137) | θ value | 4.03905 (rank : 136) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
JHD2C_MOUSE
|
||||||
| NC score | 0.011328 (rank : 138) | θ value | 4.03905 (rank : 141) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
AKAP2_MOUSE
|
||||||
| NC score | 0.011226 (rank : 139) | θ value | 1.38821 (rank : 112) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.011089 (rank : 140) | θ value | 5.27518 (rank : 156) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.011003 (rank : 141) | θ value | 0.47712 (rank : 96) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
NPDC1_HUMAN
|
||||||
| NC score | 0.010992 (rank : 142) | θ value | 4.03905 (rank : 143) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQX5, Q9BTD6, Q9BXT3, Q9NQS2, Q9Y434 | Gene names | NPDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Neural proliferation differentiation and control protein 1 precursor (NPDC-1 protein). | |||||
|
EMSY_MOUSE
|
||||||
| NC score | 0.010843 (rank : 143) | θ value | 6.88961 (rank : 158) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.010592 (rank : 144) | θ value | 5.27518 (rank : 151) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
TAB3_HUMAN
|
||||||
| NC score | 0.010330 (rank : 145) | θ value | 2.36792 (rank : 129) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
MECP2_HUMAN
|
||||||
| NC score | 0.009985 (rank : 146) | θ value | 3.0926 (rank : 133) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51608, O15233 | Gene names | MECP2 | |||
|
Domain Architecture |

|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
TIRAP_MOUSE
|
||||||
| NC score | 0.009839 (rank : 147) | θ value | 4.03905 (rank : 144) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99JY1, Q91ZW0 | Gene names | Tirap, Mal | |||
|
Domain Architecture |

|
|||||
| Description | Toll-interleukin 1 receptor domain-containing adapter protein (TIR domain-containing adapter protein) (MyD88 adapter-like protein) (Adaptor protein Wyatt). | |||||
|
SOCS6_HUMAN
|
||||||
| NC score | 0.009631 (rank : 148) | θ value | 0.813845 (rank : 105) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14544 | Gene names | SOCS6, CIS4, SOCS4 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 6 (SOCS-6) (Suppressor of cytokine signaling 4) (SOCS-4) (Cytokine-inducible SH2 protein 4) (CIS-4). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.009388 (rank : 149) | θ value | 2.36792 (rank : 124) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
SYN3_MOUSE
|
||||||
| NC score | 0.009175 (rank : 150) | θ value | 1.38821 (rank : 119) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8JZP2 | Gene names | Syn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-3 (Synapsin III). | |||||
|
CDT1_HUMAN
|
||||||
| NC score | 0.009092 (rank : 151) | θ value | 4.03905 (rank : 139) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H211, Q86XX9, Q96CJ5, Q96GK5, Q96H67, Q96HE6, Q9BWM0 | Gene names | CDT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication factor Cdt1 (Double parked homolog) (DUP). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.008859 (rank : 152) | θ value | 6.88961 (rank : 161) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CO039_HUMAN
|
||||||
| NC score | 0.008808 (rank : 153) | θ value | 3.0926 (rank : 132) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
BBX_HUMAN
|
||||||
| NC score | 0.008482 (rank : 154) | θ value | 4.03905 (rank : 138) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WY36, Q2TAJ1, Q7L3J8, Q7LBY8, Q8NDB0, Q8WY35, Q9H0J6 | Gene names | BBX, HBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
TTC28_HUMAN
|
||||||
| NC score | 0.008422 (rank : 155) | θ value | 1.81305 (rank : 122) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.008406 (rank : 156) | θ value | 0.279714 (rank : 90) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
TAB3_MOUSE
|
||||||
| NC score | 0.008347 (rank : 157) | θ value | 6.88961 (rank : 165) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
GAB1_MOUSE
|
||||||
| NC score | 0.008087 (rank : 158) | θ value | 2.36792 (rank : 126) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
UBQL3_MOUSE
|
||||||
| NC score | 0.007861 (rank : 159) | θ value | 1.81305 (rank : 123) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C5U9 | Gene names | Ubqln3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.007543 (rank : 160) | θ value | 2.36792 (rank : 128) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
BBX_MOUSE
|
||||||
| NC score | 0.006982 (rank : 161) | θ value | 5.27518 (rank : 145) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VBW5, Q3TZK1, Q6NXY8, Q6PEU3, Q8BQJ7, Q8C7E0, Q8CDQ0, Q8CDV1, Q8VI48, Q8VI49, Q8VI50, Q9CS94 | Gene names | Bbx, Hbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
DNL1_HUMAN
|
||||||
| NC score | 0.006582 (rank : 162) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
CMGA_MOUSE
|
||||||
| NC score | 0.006561 (rank : 163) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.005817 (rank : 164) | θ value | 1.81305 (rank : 121) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
JERKY_HUMAN
|
||||||
| NC score | 0.005603 (rank : 165) | θ value | 6.88961 (rank : 160) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75564, O75565 | Gene names | JRK, JH8 | |||
|
Domain Architecture |

|
|||||
| Description | Jerky protein homolog. | |||||
|
K2027_HUMAN
|
||||||
| NC score | 0.005503 (rank : 166) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.005270 (rank : 167) | θ value | 4.03905 (rank : 142) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.004890 (rank : 168) | θ value | 5.27518 (rank : 146) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.004787 (rank : 169) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.004610 (rank : 170) | θ value | 4.03905 (rank : 135) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
PHF2_MOUSE
|
||||||
| NC score | 0.004409 (rank : 171) | θ value | 5.27518 (rank : 153) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
PELI2_HUMAN
|
||||||
| NC score | 0.004338 (rank : 172) | θ value | 5.27518 (rank : 152) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HAT8 | Gene names | PELI2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein pellino homolog 2 (Pellino-2). | |||||
|
UBP8_MOUSE
|
||||||
| NC score | 0.004313 (rank : 173) | θ value | 8.99809 (rank : 177) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.004207 (rank : 174) | θ value | 5.27518 (rank : 150) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TMCC3_MOUSE
|
||||||
| NC score | 0.003973 (rank : 175) | θ value | 5.27518 (rank : 157) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R310 | Gene names | Tmcc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domains protein 3. | |||||
|
TRA2A_HUMAN
|
||||||
| NC score | 0.003450 (rank : 176) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13595 | Gene names | TRA2A | |||
|
Domain Architecture |

|
|||||
| Description | Transformer-2 protein homolog (TRA-2 alpha). | |||||
|
TRA2A_MOUSE
|
||||||
| NC score | 0.003428 (rank : 177) | θ value | 8.99809 (rank : 176) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PFR5 | Gene names | Tra2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transformer-2 protein homolog (TRA-2 alpha). | |||||
|
RBMX2_HUMAN
|
||||||
| NC score | 0.003288 (rank : 178) | θ value | 6.88961 (rank : 162) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y388, Q5JY82, Q9Y3I8 | Gene names | RBMX2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.003271 (rank : 179) | θ value | 6.88961 (rank : 159) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
EYA4_MOUSE
|
||||||
| NC score | 0.003050 (rank : 180) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z191 | Gene names | Eya4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | 0.003047 (rank : 181) | θ value | 1.38821 (rank : 113) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
RBM28_MOUSE
|
||||||
| NC score | 0.002714 (rank : 182) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CGC6, Q8BG25, Q8VEJ8, Q9CS22, Q9CSE6 | Gene names | Rbm28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 28 (RNA-binding motif protein 28). | |||||
|
ABCA2_HUMAN
|
||||||
| NC score | 0.002344 (rank : 183) | θ value | 3.0926 (rank : 130) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BZC7 | Gene names | ABCA2, ABC2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette sub-family A member 2 (ATP-binding cassette transporter 2) (ATP-binding cassette 2). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.001599 (rank : 184) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
CYTSA_MOUSE
|
||||||
| NC score | 0.000799 (rank : 185) | θ value | 4.03905 (rank : 140) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
SPEG_MOUSE
|
||||||
| NC score | -0.005796 (rank : 186) | θ value | 5.27518 (rank : 155) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||