Please be patient as the page loads
|
RECQ1_HUMAN
|
||||||
| SwissProt Accessions | P46063 | Gene names | RECQL, RECQL1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RECQ1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | P46063 | Gene names | RECQL, RECQL1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
RECQ1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995067 (rank : 2) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
BLM_MOUSE
|
||||||
| θ value | 1.94503e-103 (rank : 3) | NC score | 0.924977 (rank : 3) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O88700, O88198 | Gene names | Blm | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein homolog (EC 3.6.1.-) (mBLM). | |||||
|
BLM_HUMAN
|
||||||
| θ value | 2.15048e-102 (rank : 4) | NC score | 0.921464 (rank : 4) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 4.22625e-74 (rank : 5) | NC score | 0.867517 (rank : 6) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
WRN_HUMAN
|
||||||
| θ value | 9.43692e-66 (rank : 6) | NC score | 0.873212 (rank : 5) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
WRN_MOUSE
|
||||||
| θ value | 8.83269e-64 (rank : 7) | NC score | 0.850749 (rank : 7) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
RECQ4_HUMAN
|
||||||
| θ value | 4.2656e-42 (rank : 8) | NC score | 0.838152 (rank : 8) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
RECQ4_MOUSE
|
||||||
| θ value | 5.57106e-42 (rank : 9) | NC score | 0.827957 (rank : 9) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
DDX49_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 10) | NC score | 0.463049 (rank : 25) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 8.97725e-08 (rank : 11) | NC score | 0.463883 (rank : 24) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX17_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 12) | NC score | 0.490964 (rank : 11) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
DDX17_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 13) | NC score | 0.491039 (rank : 10) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 14) | NC score | 0.471221 (rank : 19) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX10_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 15) | NC score | 0.432554 (rank : 47) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX49_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 16) | NC score | 0.460151 (rank : 26) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX43_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 17) | NC score | 0.476080 (rank : 18) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
DDX53_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 18) | NC score | 0.451268 (rank : 33) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
DDX10_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 19) | NC score | 0.434668 (rank : 46) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX46_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 20) | NC score | 0.477536 (rank : 16) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX46_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 21) | NC score | 0.476831 (rank : 17) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
DDX23_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 22) | NC score | 0.459817 (rank : 27) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
DDX50_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 23) | NC score | 0.478902 (rank : 14) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX50_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 24) | NC score | 0.478035 (rank : 15) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX6_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 25) | NC score | 0.467106 (rank : 21) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
DDX6_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 26) | NC score | 0.466962 (rank : 22) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
DDX27_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 27) | NC score | 0.446168 (rank : 39) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX41_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 28) | NC score | 0.468412 (rank : 20) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
DDX41_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 29) | NC score | 0.466433 (rank : 23) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
DDX27_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 30) | NC score | 0.444265 (rank : 40) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX48_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 31) | NC score | 0.450384 (rank : 34) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX48_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 32) | NC score | 0.450143 (rank : 36) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
DDX47_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 33) | NC score | 0.452728 (rank : 30) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX47_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 34) | NC score | 0.451987 (rank : 31) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX5_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 35) | NC score | 0.485912 (rank : 13) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
DDX5_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 36) | NC score | 0.486501 (rank : 12) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
IF4A1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 37) | NC score | 0.435171 (rank : 44) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 38) | NC score | 0.435171 (rank : 45) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A2_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 39) | NC score | 0.436451 (rank : 42) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 40) | NC score | 0.436451 (rank : 43) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
DDX3X_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 41) | NC score | 0.450148 (rank : 35) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX3X_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 42) | NC score | 0.451321 (rank : 32) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
PL10_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 43) | NC score | 0.448690 (rank : 37) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
DDX52_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 44) | NC score | 0.454750 (rank : 28) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
SK2L2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 45) | NC score | 0.137984 (rank : 89) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P42285, Q8TAG2 | Gene names | SKIV2L2, KIAA0052 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Superkiller viralicidic activity 2-like 2 (EC 3.6.1.-) (ATP-dependent helicase SKIV2L2). | |||||
|
DDX3Y_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 46) | NC score | 0.448107 (rank : 38) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
DDX52_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 47) | NC score | 0.453932 (rank : 29) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8K301, Q8BV29 | Gene names | Ddx52, Rok1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
SK2L2_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 48) | NC score | 0.133889 (rank : 90) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9CZU3 | Gene names | Skiv2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Superkiller viralicidic activity 2-like 2 (EC 3.6.1.-) (ATP-dependent helicase SKIV2L2). | |||||
|
DDX24_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 49) | NC score | 0.429340 (rank : 53) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX24_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 50) | NC score | 0.431140 (rank : 49) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX31_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 51) | NC score | 0.429929 (rank : 52) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
DDX39_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 52) | NC score | 0.430069 (rank : 51) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O00148, Q9BVP6, Q9H5W0 | Gene names | DDX39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39) (Nuclear RNA helicase URH49). | |||||
|
DDX1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 53) | NC score | 0.413287 (rank : 59) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
DDX39_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 54) | NC score | 0.427357 (rank : 54) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8VDW0, Q3UJV4, Q8C2C2 | Gene names | Ddx39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39). | |||||
|
DDX3Y_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 55) | NC score | 0.442922 (rank : 41) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
DDX4_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 56) | NC score | 0.431947 (rank : 48) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
DDX1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 57) | NC score | 0.409925 (rank : 63) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
UAP56_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 58) | NC score | 0.420479 (rank : 57) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q13838, O43496, Q5RJ64, Q5RJ66, Q5ST94, Q5STB4, Q5STB5, Q5STB7, Q5STU4, Q5STU5, Q5STU6, Q5STU8, Q71V76 | Gene names | BAT1, UAP56 | |||
|
Domain Architecture |

|
|||||
| Description | Spliceosome RNA helicase BAT1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (ATP-dependent RNA helicase p47) (HLA-B-associated transcript-1). | |||||
|
UAP56_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 59) | NC score | 0.420405 (rank : 58) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Z1N5, Q8HW97 | Gene names | Bat1, Bat1a, Uap56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spliceosome RNA helicase Bat1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (HLA-B-associated transcript 1). | |||||
|
DDX51_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 60) | NC score | 0.412557 (rank : 60) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
SKIV2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 61) | NC score | 0.143147 (rank : 88) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15477, O15005, Q12902, Q15476 | Gene names | SKIV2L, DDX13, SKI2W, SKIV2, W | |||
|
Domain Architecture |

|
|||||
| Description | Helicase SKI2W (EC 3.6.1.-) (Helicase-like protein) (HLP). | |||||
|
DDX54_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 62) | NC score | 0.423293 (rank : 56) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
DDX28_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 63) | NC score | 0.410869 (rank : 61) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX51_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 64) | NC score | 0.410588 (rank : 62) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX54_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 65) | NC score | 0.426739 (rank : 55) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
U520_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 66) | NC score | 0.189869 (rank : 80) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75643, O94884, Q6PX59, Q96IF2, Q9H7S0 | Gene names | ASCC3L1, HELIC2, KIAA0788 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U5 small nuclear ribonucleoprotein 200 kDa helicase (EC 3.6.1.-) (U5 snRNP-specific 200 kDa protein) (U5-200KD) (Activating signal cointegrator 1 complex subunit 3-like 1) (BRR2 homolog). | |||||
|
DDX28_HUMAN
|
||||||
| θ value | 0.163984 (rank : 67) | NC score | 0.397608 (rank : 64) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 68) | NC score | 0.430267 (rank : 50) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
DICER_MOUSE
|
||||||
| θ value | 0.47712 (rank : 69) | NC score | 0.208175 (rank : 78) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
DICER_HUMAN
|
||||||
| θ value | 0.62314 (rank : 70) | NC score | 0.206005 (rank : 79) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
VDP_HUMAN
|
||||||
| θ value | 0.62314 (rank : 71) | NC score | 0.028083 (rank : 101) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60763 | Gene names | VDP | |||
|
Domain Architecture |

|
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 72) | NC score | 0.010600 (rank : 109) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
SDCG1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 73) | NC score | 0.040493 (rank : 96) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60524, Q8WW70, Q9NWG1 | Gene names | SDCCAG1 | |||
|
Domain Architecture |
|
|||||
| Description | Serologically defined colon cancer antigen 1 (Antigen NY-CO-1). | |||||
|
ZN365_MOUSE
|
||||||
| θ value | 0.813845 (rank : 74) | NC score | 0.033596 (rank : 97) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BG89, Q80TQ4, Q8BK39, Q8BXT2 | Gene names | Znf365, Kiaa0844, Zfp365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF365 (Su48). | |||||
|
TACC3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 75) | NC score | 0.012876 (rank : 107) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
LGP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.179384 (rank : 83) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
TPM1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.009892 (rank : 110) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P58771, P02558, P19354, P46902, P99034 | Gene names | Tpm1, Tpm-1, Tpma | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
IFIH1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.172933 (rank : 85) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
TPM1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.009689 (rank : 113) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P09493, P09494, P10469, Q86W64, Q96IK2, Q9UCY9 | Gene names | TPM1, TMSA | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
DDX20_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.393442 (rank : 65) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.014353 (rank : 106) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
PROF2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.023560 (rank : 102) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35080, Q8WVF9, Q9HBK2 | Gene names | PFN2 | |||
|
Domain Architecture |
|
|||||
| Description | Profilin-2 (Profilin II). | |||||
|
ZN365_HUMAN
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.029308 (rank : 100) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q70YC5, O94930, Q5HYE6, Q68SG8, Q6NSK2, Q6P9D4, Q70YC6, Q70YC7 | Gene names | ZNF365, KIAA0844 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF365. | |||||
|
HELC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.150190 (rank : 87) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
IFIH1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.175078 (rank : 84) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.009760 (rank : 111) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.011495 (rank : 108) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
PROF2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.022397 (rank : 103) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJV2, Q9ES49, Q9ES50 | Gene names | Pfn2 | |||
|
Domain Architecture |
|
|||||
| Description | Profilin-2 (Profilin II). | |||||
|
SDCG1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.032602 (rank : 98) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CCP0, Q66JX6, Q8C9R6, Q8CA65, Q8JZT9, Q8R072, Q9CW30, Q9CYB8, Q9D4A9 | Gene names | Sdccag1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1. | |||||
|
CP250_MOUSE
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.003364 (rank : 115) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
DDX20_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.389293 (rank : 66) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DDX55_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.379342 (rank : 68) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
FA61A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.016363 (rank : 104) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8ND56, Q76LX7, Q96AR3, Q96K73, Q96SN5, Q9UFR3 | Gene names | FAM61A, C19orf13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM61A (Putative alpha synuclein-binding protein) (AlphaSNBP). | |||||
|
FA61A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.016220 (rank : 105) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K2F8, Q9CTG8 | Gene names | Fam61a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM61A. | |||||
|
FINC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.003030 (rank : 116) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
K0562_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.009722 (rank : 112) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60308, Q5JSQ3, Q5SR24, Q5SR25, Q6PKF5, Q86W32, Q86X14 | Gene names | KIAA0562 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0562. | |||||
|
LGP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.170800 (rank : 86) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
VDP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.030597 (rank : 99) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z1Z0, Q3T9L9, Q3TH58, Q3U1C7, Q3UMW6, Q91WE7, Q99JZ5 | Gene names | Vdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.002995 (rank : 117) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
NUF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.004278 (rank : 114) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99P69, Q8BTK7, Q8VE05, Q9CST5 | Gene names | Cdca1, Nuf2r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (Cell division cycle-associated protein 1). | |||||
|
CC026_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.066351 (rank : 95) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BQ75 | Gene names | C3orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26. | |||||
|
DD19A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.364654 (rank : 71) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.363352 (rank : 73) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DD19B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.363797 (rank : 72) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
DDX18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.387315 (rank : 67) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
DDX18_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.379062 (rank : 69) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
DDX25_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.361633 (rank : 74) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX25_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.358187 (rank : 75) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX55_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.375998 (rank : 70) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX56_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.356187 (rank : 77) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NY93, Q9H9I8 | Gene names | DDX56, DDX21, NOH61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase) (DEAD-box protein 21). | |||||
|
DDX56_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.356742 (rank : 76) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
DDX58_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.084626 (rank : 92) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
DDX58_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.098010 (rank : 91) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
EXDL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.072924 (rank : 93) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NVH0, Q8N3D3 | Gene names | EXDL2, C14orf114 | |||
|
Domain Architecture |
|
|||||
| Description | Exonuclease 3'-5' domain-like-containing protein 2. | |||||
|
EXDL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.070633 (rank : 94) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VEG4 | Gene names | Exdl2 | |||
|
Domain Architecture |
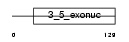
|
|||||
| Description | Exonuclease 3'-5' domain-like-containing protein 2. | |||||
|
FANCM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.185596 (rank : 81) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
FANCM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.184901 (rank : 82) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
RECQ1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | P46063 | Gene names | RECQL, RECQL1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
RECQ1_MOUSE
|
||||||
| NC score | 0.995067 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
BLM_MOUSE
|
||||||
| NC score | 0.924977 (rank : 3) | θ value | 1.94503e-103 (rank : 3) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O88700, O88198 | Gene names | Blm | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein homolog (EC 3.6.1.-) (mBLM). | |||||
|
BLM_HUMAN
|
||||||
| NC score | 0.921464 (rank : 4) | θ value | 2.15048e-102 (rank : 4) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
WRN_HUMAN
|
||||||
| NC score | 0.873212 (rank : 5) | θ value | 9.43692e-66 (rank : 6) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.867517 (rank : 6) | θ value | 4.22625e-74 (rank : 5) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
WRN_MOUSE
|
||||||
| NC score | 0.850749 (rank : 7) | θ value | 8.83269e-64 (rank : 7) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
RECQ4_HUMAN
|
||||||
| NC score | 0.838152 (rank : 8) | θ value | 4.2656e-42 (rank : 8) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
RECQ4_MOUSE
|
||||||
| NC score | 0.827957 (rank : 9) | θ value | 5.57106e-42 (rank : 9) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
DDX17_MOUSE
|
||||||
| NC score | 0.491039 (rank : 10) | θ value | 1.17247e-07 (rank : 13) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
DDX17_HUMAN
|
||||||
| NC score | 0.490964 (rank : 11) | θ value | 1.17247e-07 (rank : 12) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
DDX5_MOUSE
|
||||||
| NC score | 0.486501 (rank : 12) | θ value | 5.44631e-05 (rank : 36) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
DDX5_HUMAN
|
||||||
| NC score | 0.485912 (rank : 13) | θ value | 5.44631e-05 (rank : 35) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
DDX50_HUMAN
|
||||||
| NC score | 0.478902 (rank : 14) | θ value | 2.21117e-06 (rank : 23) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX50_MOUSE
|
||||||
| NC score | 0.478035 (rank : 15) | θ value | 2.88788e-06 (rank : 24) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX46_HUMAN
|
||||||
| NC score | 0.477536 (rank : 16) | θ value | 1.29631e-06 (rank : 20) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX46_MOUSE
|
||||||
| NC score | 0.476831 (rank : 17) | θ value | 1.29631e-06 (rank : 21) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
DDX43_HUMAN
|
||||||
| NC score | 0.476080 (rank : 18) | θ value | 9.92553e-07 (rank : 17) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.471221 (rank : 19) | θ value | 1.17247e-07 (rank : 14) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX41_HUMAN
|
||||||
| NC score | 0.468412 (rank : 20) | θ value | 1.09739e-05 (rank : 28) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
DDX6_HUMAN
|
||||||
| NC score | 0.467106 (rank : 21) | θ value | 4.92598e-06 (rank : 25) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
DDX6_MOUSE
|
||||||
| NC score | 0.466962 (rank : 22) | θ value | 4.92598e-06 (rank : 26) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
DDX41_MOUSE
|
||||||
| NC score | 0.466433 (rank : 23) | θ value | 1.09739e-05 (rank : 29) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.463883 (rank : 24) | θ value | 8.97725e-08 (rank : 11) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX49_MOUSE
|
||||||
| NC score | 0.463049 (rank : 25) | θ value | 6.87365e-08 (rank : 10) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX49_HUMAN
|
||||||
| NC score | 0.460151 (rank : 26) | θ value | 7.59969e-07 (rank : 16) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX23_HUMAN
|
||||||
| NC score | 0.459817 (rank : 27) | θ value | 2.21117e-06 (rank : 22) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
DDX52_HUMAN
|
||||||
| NC score | 0.454750 (rank : 28) | θ value | 0.00228821 (rank : 44) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX52_MOUSE
|
||||||
| NC score | 0.453932 (rank : 29) | θ value | 0.00665767 (rank : 47) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8K301, Q8BV29 | Gene names | Ddx52, Rok1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX47_HUMAN
|
||||||
| NC score | 0.452728 (rank : 30) | θ value | 1.87187e-05 (rank : 33) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX47_MOUSE
|
||||||
| NC score | 0.451987 (rank : 31) | θ value | 2.44474e-05 (rank : 34) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX3X_MOUSE
|
||||||
| NC score | 0.451321 (rank : 32) | θ value | 0.000786445 (rank : 42) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
DDX53_HUMAN
|
||||||
| NC score | 0.451268 (rank : 33) | θ value | 9.92553e-07 (rank : 18) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
DDX48_HUMAN
|
||||||
| NC score | 0.450384 (rank : 34) | θ value | 1.43324e-05 (rank : 31) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX3X_HUMAN
|
||||||
| NC score | 0.450148 (rank : 35) | θ value | 0.000786445 (rank : 41) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX48_MOUSE
|
||||||
| NC score | 0.450143 (rank : 36) | θ value | 1.43324e-05 (rank : 32) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
PL10_MOUSE
|
||||||
| NC score | 0.448690 (rank : 37) | θ value | 0.00134147 (rank : 43) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
DDX3Y_HUMAN
|
||||||
| NC score | 0.448107 (rank : 38) | θ value | 0.00509761 (rank : 46) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
DDX27_MOUSE
|
||||||
| NC score | 0.446168 (rank : 39) | θ value | 1.09739e-05 (rank : 27) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX27_HUMAN
|
||||||
| NC score | 0.444265 (rank : 40) | θ value | 1.43324e-05 (rank : 30) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX3Y_MOUSE
|
||||||
| NC score | 0.442922 (rank : 41) | θ value | 0.0193708 (rank : 55) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
IF4A2_HUMAN
|
||||||
| NC score | 0.436451 (rank : 42) | θ value | 0.00020696 (rank : 39) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| NC score | 0.436451 (rank : 43) | θ value | 0.00020696 (rank : 40) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A1_HUMAN
|
||||||
| NC score | 0.435171 (rank : 44) | θ value | 0.00020696 (rank : 37) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| NC score | 0.435171 (rank : 45) | θ value | 0.00020696 (rank : 38) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
DDX10_HUMAN
|
||||||
| NC score | 0.434668 (rank : 46) | θ value | 1.29631e-06 (rank : 19) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX10_MOUSE
|
||||||
| NC score | 0.432554 (rank : 47) | θ value | 2.61198e-07 (rank : 15) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX4_HUMAN
|
||||||
| NC score | 0.431947 (rank : 48) | θ value | 0.0193708 (rank : 56) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
DDX24_MOUSE
|
||||||
| NC score | 0.431140 (rank : 49) | θ value | 0.00869519 (rank : 50) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX4_MOUSE
|
||||||
| NC score | 0.430267 (rank : 50) | θ value | 0.163984 (rank : 68) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
DDX39_HUMAN
|
||||||
| NC score | 0.430069 (rank : 51) | θ value | 0.0113563 (rank : 52) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O00148, Q9BVP6, Q9H5W0 | Gene names | DDX39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39) (Nuclear RNA helicase URH49). | |||||
|
DDX31_HUMAN
|
||||||
| NC score | 0.429929 (rank : 52) | θ value | 0.0113563 (rank : 51) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
DDX24_HUMAN
|
||||||
| NC score | 0.429340 (rank : 53) | θ value | 0.00869519 (rank : 49) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX39_MOUSE
|
||||||
| NC score | 0.427357 (rank : 54) | θ value | 0.0148317 (rank : 54) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8VDW0, Q3UJV4, Q8C2C2 | Gene names | Ddx39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39). | |||||
|
DDX54_MOUSE
|
||||||
| NC score | 0.426739 (rank : 55) | θ value | 0.0961366 (rank : 65) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
DDX54_HUMAN
|
||||||
| NC score | 0.423293 (rank : 56) | θ value | 0.0563607 (rank : 62) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
UAP56_HUMAN
|
||||||
| NC score | 0.420479 (rank : 57) | θ value | 0.0330416 (rank : 58) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q13838, O43496, Q5RJ64, Q5RJ66, Q5ST94, Q5STB4, Q5STB5, Q5STB7, Q5STU4, Q5STU5, Q5STU6, Q5STU8, Q71V76 | Gene names | BAT1, UAP56 | |||
|
Domain Architecture |

|
|||||
| Description | Spliceosome RNA helicase BAT1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (ATP-dependent RNA helicase p47) (HLA-B-associated transcript-1). | |||||
|
UAP56_MOUSE
|
||||||
| NC score | 0.420405 (rank : 58) | θ value | 0.0330416 (rank : 59) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Z1N5, Q8HW97 | Gene names | Bat1, Bat1a, Uap56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spliceosome RNA helicase Bat1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (HLA-B-associated transcript 1). | |||||
|
DDX1_MOUSE
|
||||||
| NC score | 0.413287 (rank : 59) | θ value | 0.0148317 (rank : 53) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
DDX51_HUMAN
|
||||||
| NC score | 0.412557 (rank : 60) | θ value | 0.0431538 (rank : 60) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX28_MOUSE
|
||||||
| NC score | 0.410869 (rank : 61) | θ value | 0.0961366 (rank : 63) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX51_MOUSE
|
||||||
| NC score | 0.410588 (rank : 62) | θ value | 0.0961366 (rank : 64) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX1_HUMAN
|
||||||
| NC score | 0.409925 (rank : 63) | θ value | 0.0330416 (rank : 57) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
DDX28_HUMAN
|
||||||
| NC score | 0.397608 (rank : 64) | θ value | 0.163984 (rank : 67) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX20_HUMAN
|
||||||
| NC score | 0.393442 (rank : 65) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
DDX20_MOUSE
|
||||||
| NC score | 0.389293 (rank : 66) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DDX18_HUMAN
|
||||||
| NC score | 0.387315 (rank : 67) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
DDX55_MOUSE
|
||||||
| NC score | 0.379342 (rank : 68) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX18_MOUSE
|
||||||
| NC score | 0.379062 (rank : 69) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
DDX55_HUMAN
|
||||||
| NC score | 0.375998 (rank : 70) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DD19A_HUMAN
|
||||||
| NC score | 0.364654 (rank : 71) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19B_HUMAN
|
||||||
| NC score | 0.363797 (rank : 72) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
DD19A_MOUSE
|
||||||
| NC score | 0.363352 (rank : 73) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DDX25_HUMAN
|
||||||
| NC score | 0.361633 (rank : 74) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX25_MOUSE
|
||||||
| NC score | 0.358187 (rank : 75) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX56_MOUSE
|
||||||
| NC score | 0.356742 (rank : 76) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
DDX56_HUMAN
|
||||||
| NC score | 0.356187 (rank : 77) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NY93, Q9H9I8 | Gene names | DDX56, DDX21, NOH61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase) (DEAD-box protein 21). | |||||
|
DICER_MOUSE
|
||||||
| NC score | 0.208175 (rank : 78) | θ value | 0.47712 (rank : 69) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
DICER_HUMAN
|
||||||
| NC score | 0.206005 (rank : 79) | θ value | 0.62314 (rank : 70) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
U520_HUMAN
|
||||||
| NC score | 0.189869 (rank : 80) | θ value | 0.0961366 (rank : 66) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75643, O94884, Q6PX59, Q96IF2, Q9H7S0 | Gene names | ASCC3L1, HELIC2, KIAA0788 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U5 small nuclear ribonucleoprotein 200 kDa helicase (EC 3.6.1.-) (U5 snRNP-specific 200 kDa protein) (U5-200KD) (Activating signal cointegrator 1 complex subunit 3-like 1) (BRR2 homolog). | |||||
|
FANCM_HUMAN
|
||||||
| NC score | 0.185596 (rank : 81) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
FANCM_MOUSE
|
||||||
| NC score | 0.184901 (rank : 82) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
LGP2_HUMAN
|
||||||
| NC score | 0.179384 (rank : 83) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
IFIH1_HUMAN
|
||||||
| NC score | 0.175078 (rank : 84) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
IFIH1_MOUSE
|
||||||
| NC score | 0.172933 (rank : 85) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
LGP2_MOUSE
|
||||||
| NC score | 0.170800 (rank : 86) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
HELC1_HUMAN
|
||||||
| NC score | 0.150190 (rank : 87) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
SKIV2_HUMAN
|
||||||
| NC score | 0.143147 (rank : 88) | θ value | 0.0431538 (rank : 61) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15477, O15005, Q12902, Q15476 | Gene names | SKIV2L, DDX13, SKI2W, SKIV2, W | |||
|
Domain Architecture |

|
|||||
| Description | Helicase SKI2W (EC 3.6.1.-) (Helicase-like protein) (HLP). | |||||
|
SK2L2_HUMAN
|
||||||
| NC score | 0.137984 (rank : 89) | θ value | 0.00298849 (rank : 45) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P42285, Q8TAG2 | Gene names | SKIV2L2, KIAA0052 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Superkiller viralicidic activity 2-like 2 (EC 3.6.1.-) (ATP-dependent helicase SKIV2L2). | |||||
|
SK2L2_MOUSE
|
||||||
| NC score | 0.133889 (rank : 90) | θ value | 0.00665767 (rank : 48) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9CZU3 | Gene names | Skiv2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Superkiller viralicidic activity 2-like 2 (EC 3.6.1.-) (ATP-dependent helicase SKIV2L2). | |||||
|
DDX58_MOUSE
|
||||||
| NC score | 0.098010 (rank : 91) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
DDX58_HUMAN
|
||||||
| NC score | 0.084626 (rank : 92) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
EXDL2_HUMAN
|
||||||
| NC score | 0.072924 (rank : 93) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NVH0, Q8N3D3 | Gene names | EXDL2, C14orf114 | |||
|
Domain Architecture |
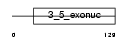
|
|||||
| Description | Exonuclease 3'-5' domain-like-containing protein 2. | |||||
|
EXDL2_MOUSE
|
||||||
| NC score | 0.070633 (rank : 94) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VEG4 | Gene names | Exdl2 | |||
|
Domain Architecture |
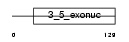
|
|||||
| Description | Exonuclease 3'-5' domain-like-containing protein 2. | |||||
|
CC026_HUMAN
|
||||||
| NC score | 0.066351 (rank : 95) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BQ75 | Gene names | C3orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26. | |||||
|
SDCG1_HUMAN
|
||||||
| NC score | 0.040493 (rank : 96) | θ value | 0.813845 (rank : 73) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60524, Q8WW70, Q9NWG1 | Gene names | SDCCAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1 (Antigen NY-CO-1). | |||||
|
ZN365_MOUSE
|
||||||
| NC score | 0.033596 (rank : 97) | θ value | 0.813845 (rank : 74) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BG89, Q80TQ4, Q8BK39, Q8BXT2 | Gene names | Znf365, Kiaa0844, Zfp365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF365 (Su48). | |||||
|
SDCG1_MOUSE
|
||||||
| NC score | 0.032602 (rank : 98) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CCP0, Q66JX6, Q8C9R6, Q8CA65, Q8JZT9, Q8R072, Q9CW30, Q9CYB8, Q9D4A9 | Gene names | Sdccag1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1. | |||||
|
VDP_MOUSE
|
||||||
| NC score | 0.030597 (rank : 99) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z1Z0, Q3T9L9, Q3TH58, Q3U1C7, Q3UMW6, Q91WE7, Q99JZ5 | Gene names | Vdp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
ZN365_HUMAN
|
||||||
| NC score | 0.029308 (rank : 100) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q70YC5, O94930, Q5HYE6, Q68SG8, Q6NSK2, Q6P9D4, Q70YC6, Q70YC7 | Gene names | ZNF365, KIAA0844 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF365. | |||||
|
VDP_HUMAN
|
||||||
| NC score | 0.028083 (rank : 101) | θ value | 0.62314 (rank : 71) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60763 | Gene names | VDP | |||
|
Domain Architecture |

|
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
PROF2_HUMAN
|
||||||
| NC score | 0.023560 (rank : 102) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35080, Q8WVF9, Q9HBK2 | Gene names | PFN2 | |||
|
Domain Architecture |

|
|||||
| Description | Profilin-2 (Profilin II). | |||||
|
PROF2_MOUSE
|
||||||
| NC score | 0.022397 (rank : 103) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJV2, Q9ES49, Q9ES50 | Gene names | Pfn2 | |||
|
Domain Architecture |

|
|||||
| Description | Profilin-2 (Profilin II). | |||||
|
FA61A_HUMAN
|
||||||
| NC score | 0.016363 (rank : 104) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8ND56, Q76LX7, Q96AR3, Q96K73, Q96SN5, Q9UFR3 | Gene names | FAM61A, C19orf13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM61A (Putative alpha synuclein-binding protein) (AlphaSNBP). | |||||
|
FA61A_MOUSE
|
||||||
| NC score | 0.016220 (rank : 105) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K2F8, Q9CTG8 | Gene names | Fam61a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM61A. | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.014353 (rank : 106) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
TACC3_HUMAN
|
||||||
| NC score | 0.012876 (rank : 107) | θ value | 1.06291 (rank : 75) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.011495 (rank : 108) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.010600 (rank : 109) | θ value | 0.813845 (rank : 72) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
TPM1_MOUSE
|
||||||
| NC score | 0.009892 (rank : 110) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P58771, P02558, P19354, P46902, P99034 | Gene names | Tpm1, Tpm-1, Tpma | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.009760 (rank : 111) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
K0562_HUMAN
|
||||||
| NC score | 0.009722 (rank : 112) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60308, Q5JSQ3, Q5SR24, Q5SR25, Q6PKF5, Q86W32, Q86X14 | Gene names | KIAA0562 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0562. | |||||
|
TPM1_HUMAN
|
||||||
| NC score | 0.009689 (rank : 113) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P09493, P09494, P10469, Q86W64, Q96IK2, Q9UCY9 | Gene names | TPM1, TMSA | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
NUF2_MOUSE
|
||||||
| NC score | 0.004278 (rank : 114) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99P69, Q8BTK7, Q8VE05, Q9CST5 | Gene names | Cdca1, Nuf2r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (Cell division cycle-associated protein 1). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.003364 (rank : 115) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
FINC_MOUSE
|
||||||
| NC score | 0.003030 (rank : 116) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.002995 (rank : 117) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||