Please be patient as the page loads
|
NPM_HUMAN
|
||||||
| SwissProt Accessions | P06748, P08693, Q12826, Q13440, Q13441, Q14115, Q5EU94, Q5EU95, Q5EU96, Q5EU97, Q5EU98, Q5EU99, Q6V962, Q8WTW5, Q96AT6, Q96DC4, Q96EA5 | Gene names | NPM1, NPM | |||
|
Domain Architecture |

|
|||||
| Description | Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NPM_HUMAN
|
||||||
| θ value | 4.29305e-135 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P06748, P08693, Q12826, Q13440, Q13441, Q14115, Q5EU94, Q5EU95, Q5EU96, Q5EU97, Q5EU98, Q5EU99, Q6V962, Q8WTW5, Q96AT6, Q96DC4, Q96EA5 | Gene names | NPM1, NPM | |||
|
Domain Architecture |

|
|||||
| Description | Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38). | |||||
|
NPM_MOUSE
|
||||||
| θ value | 1.5801e-129 (rank : 2) | NC score | 0.969446 (rank : 2) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61937 | Gene names | Npm1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38). | |||||
|
NPM3_HUMAN
|
||||||
| θ value | 1.02238e-19 (rank : 3) | NC score | 0.777444 (rank : 3) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75607, Q9UNY6 | Gene names | NPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoplasmin-3. | |||||
|
NPM3_MOUSE
|
||||||
| θ value | 3.28887e-18 (rank : 4) | NC score | 0.771594 (rank : 4) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CPP0, Q3UJ58 | Gene names | Npm3 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoplasmin-3. | |||||
|
NPM2_MOUSE
|
||||||
| θ value | 1.58096e-12 (rank : 5) | NC score | 0.677427 (rank : 5) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80W85, Q8BW23 | Gene names | Npm2 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoplasmin-2. | |||||
|
NPM2_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 6) | NC score | 0.655664 (rank : 6) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86SE8 | Gene names | NPM2 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoplasmin-2. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 7) | NC score | 0.104466 (rank : 7) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 8) | NC score | 0.097564 (rank : 9) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 9) | NC score | 0.087436 (rank : 16) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.087676 (rank : 15) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
H14_HUMAN
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.084063 (rank : 19) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
LRMP_MOUSE
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.057018 (rank : 37) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60664, Q7TMU8 | Gene names | Lrmp, Jaw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphoid-restricted membrane protein (Protein Jaw1). | |||||
|
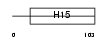
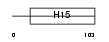
H15_HUMAN
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.095971 (rank : 10) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P16401, Q14529 | Gene names | HIST1H1B, H1F5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (Histone H1a). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.102611 (rank : 8) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.092992 (rank : 11) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
ZCPW1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.071458 (rank : 26) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
ENL_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.087911 (rank : 14) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.049954 (rank : 56) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.082785 (rank : 20) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.055996 (rank : 40) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
VPS72_MOUSE
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.091220 (rank : 13) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
ZC313_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.067548 (rank : 29) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
CCD55_MOUSE
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.060727 (rank : 34) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.087356 (rank : 17) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
NFH_HUMAN
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.039690 (rank : 63) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PARG_HUMAN
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.056484 (rank : 39) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86W56, Q6E4P6, Q6E4P7, Q7Z742, Q9Y4W7 | Gene names | PARG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
DDX20_MOUSE
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.021799 (rank : 74) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.079693 (rank : 23) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
H10_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.091506 (rank : 12) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P07305 | Gene names | H1F0, H1FV | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.0 (H1(0)) (Histone H1'). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.058749 (rank : 35) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
VPS72_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.084529 (rank : 18) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
H14_MOUSE
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.073033 (rank : 24) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.040068 (rank : 62) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.062046 (rank : 33) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.027948 (rank : 68) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
RBM19_MOUSE
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.025907 (rank : 72) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R3C6, Q8BHR0, Q9CW63 | Gene names | Rbm19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
ARI5B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.040296 (rank : 61) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.081516 (rank : 22) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
DDX20_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.017042 (rank : 82) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
IF4B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.052860 (rank : 49) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P23588 | Gene names | EIF4B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.034163 (rank : 65) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
ZN592_MOUSE
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.007934 (rank : 85) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BHZ4, Q80XM1 | Gene names | Znf592, Kiaa0211, Zfp592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 592 (Zfp-592). | |||||
|
CASZ1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.017642 (rank : 81) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 473 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86V15, Q5T9S1, Q8WX49, Q8WX50, Q9BT16, Q9NXC6 | Gene names | CASZ1, CST, ZNF693 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Castor homolog 1 zinc finger protein (Castor-related protein) (Zinc finger protein 693). | |||||
|
GCX1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.018315 (rank : 77) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96NM4, Q5TE33, Q5TE34, Q5TE35, Q96IC9, Q9BQN5 | Gene names | GCX1, C20orf100 | |||
|
Domain Architecture |

|
|||||
| Description | Granulosa cell HMG box protein 1 (GCX-1). | |||||
|
H10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.070834 (rank : 27) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10922 | Gene names | H1f0, H1fv | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1' (H1.0) (H1(0)). | |||||
|
IF4B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.045272 (rank : 59) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.025979 (rank : 71) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
NOL8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.047066 (rank : 57) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.026038 (rank : 70) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
REXO1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.031102 (rank : 66) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TT28, Q3UMP1, Q69ZR0, Q6NSQ6, Q6PI95, Q9DA29 | Gene names | Rexo1, Kiaa1138, Tceb3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.016133 (rank : 83) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
B3A3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.018046 (rank : 79) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48751 | Gene names | SLC4A3, AE3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3) (Cardiac/brain band 3-like protein) (CAE3/BAE3). | |||||
|
BRDT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.019348 (rank : 76) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
H13_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.055115 (rank : 43) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P16402 | Gene names | HIST1H1D, H1F3 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.3 (Histone H1c). | |||||
|
HMGA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.035164 (rank : 64) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P52926 | Gene names | HMGA2, HMGIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein HMGI-C (High mobility group AT-hook protein 2). | |||||
|
OXR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.044789 (rank : 60) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.023327 (rank : 73) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
THOC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.072885 (rank : 25) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
TOP2A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.029492 (rank : 67) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q01320 | Gene names | Top2a, Top-2, Top2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.016018 (rank : 84) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
ZCH11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.027777 (rank : 69) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.020452 (rank : 75) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
B3A3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.017868 (rank : 80) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P16283 | Gene names | Slc4a3, Ae3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3). | |||||
|
FOG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.001943 (rank : 86) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
H13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.052497 (rank : 50) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P43277, Q8C6M4 | Gene names | Hist1h1d, H1f3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (H1 VAR.4) (H1d). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.046344 (rank : 58) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.018236 (rank : 78) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.053421 (rank : 47) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.082416 (rank : 21) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
AF9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.055657 (rank : 41) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.053827 (rank : 45) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.055171 (rank : 42) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.065561 (rank : 31) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.063652 (rank : 32) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.050654 (rank : 54) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
H11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.052353 (rank : 51) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P43275 | Gene names | Hist1h1a, H1f1 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.1 (H1 VAR.3) (H1a). | |||||
|
H15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.053489 (rank : 46) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P43276, Q9CRM8 | Gene names | Hist1h1b, H1f5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (H1 VAR.5) (H1b). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.056767 (rank : 38) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.069153 (rank : 28) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
NUCKS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.066713 (rank : 30) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.053121 (rank : 48) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.054403 (rank : 44) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
RSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.050897 (rank : 53) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SFR11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.058102 (rank : 36) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
T2FA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.050499 (rank : 55) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.051488 (rank : 52) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
NPM_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.29305e-135 (rank : 1) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P06748, P08693, Q12826, Q13440, Q13441, Q14115, Q5EU94, Q5EU95, Q5EU96, Q5EU97, Q5EU98, Q5EU99, Q6V962, Q8WTW5, Q96AT6, Q96DC4, Q96EA5 | Gene names | NPM1, NPM | |||
|
Domain Architecture |

|
|||||
| Description | Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38). | |||||
|
NPM_MOUSE
|
||||||
| NC score | 0.969446 (rank : 2) | θ value | 1.5801e-129 (rank : 2) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61937 | Gene names | Npm1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38). | |||||
|
NPM3_HUMAN
|
||||||
| NC score | 0.777444 (rank : 3) | θ value | 1.02238e-19 (rank : 3) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75607, Q9UNY6 | Gene names | NPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoplasmin-3. | |||||
|
NPM3_MOUSE
|
||||||
| NC score | 0.771594 (rank : 4) | θ value | 3.28887e-18 (rank : 4) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CPP0, Q3UJ58 | Gene names | Npm3 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoplasmin-3. | |||||
|
NPM2_MOUSE
|
||||||
| NC score | 0.677427 (rank : 5) | θ value | 1.58096e-12 (rank : 5) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80W85, Q8BW23 | Gene names | Npm2 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoplasmin-2. | |||||
|
NPM2_HUMAN
|
||||||
| NC score | 0.655664 (rank : 6) | θ value | 3.89403e-11 (rank : 6) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86SE8 | Gene names | NPM2 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoplasmin-2. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.104466 (rank : 7) | θ value | 0.00869519 (rank : 7) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.102611 (rank : 8) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.097564 (rank : 9) | θ value | 0.0563607 (rank : 8) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
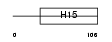
H15_HUMAN
|
||||||
| NC score | 0.095971 (rank : 10) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P16401, Q14529 | Gene names | HIST1H1B, H1F5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (Histone H1a). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.092992 (rank : 11) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
H10_HUMAN
|
||||||
| NC score | 0.091506 (rank : 12) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P07305 | Gene names | H1F0, H1FV | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.0 (H1(0)) (Histone H1'). | |||||
|
VPS72_MOUSE
|
||||||
| NC score | 0.091220 (rank : 13) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.087911 (rank : 14) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.087676 (rank : 15) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.087436 (rank : 16) | θ value | 0.0563607 (rank : 9) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.087356 (rank : 17) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
VPS72_HUMAN
|
||||||
| NC score | 0.084529 (rank : 18) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
H14_HUMAN
|
||||||
| NC score | 0.084063 (rank : 19) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.082785 (rank : 20) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.082416 (rank : 21) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.081516 (rank : 22) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.079693 (rank : 23) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
H14_MOUSE
|
||||||
| NC score | 0.073033 (rank : 24) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.072885 (rank : 25) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
ZCPW1_MOUSE
|
||||||
| NC score | 0.071458 (rank : 26) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
H10_MOUSE
|
||||||
| NC score | 0.070834 (rank : 27) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10922 | Gene names | H1f0, H1fv | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1' (H1.0) (H1(0)). | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.069153 (rank : 28) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.067548 (rank : 29) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
NUCKS_MOUSE
|
||||||
| NC score | 0.066713 (rank : 30) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.065561 (rank : 31) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.063652 (rank : 32) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.062046 (rank : 33) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.060727 (rank : 34) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.058749 (rank : 35) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
SFR11_HUMAN
|
||||||
| NC score | 0.058102 (rank : 36) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
LRMP_MOUSE
|
||||||
| NC score | 0.057018 (rank : 37) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60664, Q7TMU8 | Gene names | Lrmp, Jaw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphoid-restricted membrane protein (Protein Jaw1). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.056767 (rank : 38) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
PARG_HUMAN
|
||||||
| NC score | 0.056484 (rank : 39) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86W56, Q6E4P6, Q6E4P7, Q7Z742, Q9Y4W7 | Gene names | PARG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.055996 (rank : 40) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
AF9_HUMAN
|
||||||
| NC score | 0.055657 (rank : 41) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.055171 (rank : 42) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
H13_HUMAN
|
||||||
| NC score | 0.055115 (rank : 43) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P16402 | Gene names | HIST1H1D, H1F3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (Histone H1c). | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.054403 (rank : 44) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.053827 (rank : 45) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
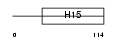
H15_MOUSE
|
||||||
| NC score | 0.053489 (rank : 46) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P43276, Q9CRM8 | Gene names | Hist1h1b, H1f5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (H1 VAR.5) (H1b). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.053421 (rank : 47) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.053121 (rank : 48) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
IF4B_HUMAN
|
||||||
| NC score | 0.052860 (rank : 49) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P23588 | Gene names | EIF4B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
H13_MOUSE
|
||||||
| NC score | 0.052497 (rank : 50) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P43277, Q8C6M4 | Gene names | Hist1h1d, H1f3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (H1 VAR.4) (H1d). | |||||
|
H11_MOUSE
|
||||||
| NC score | 0.052353 (rank : 51) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P43275 | Gene names | Hist1h1a, H1f1 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.1 (H1 VAR.3) (H1a). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.051488 (rank : 52) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.050897 (rank : 53) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.050654 (rank : 54) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.050499 (rank : 55) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.049954 (rank : 56) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
NOL8_HUMAN
|
||||||
| NC score | 0.047066 (rank : 57) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.046344 (rank : 58) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
IF4B_MOUSE
|
||||||
| NC score | 0.045272 (rank : 59) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
OXR1_MOUSE
|
||||||
| NC score | 0.044789 (rank : 60) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
ARI5B_HUMAN
|
||||||
| NC score | 0.040296 (rank : 61) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.040068 (rank : 62) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.039690 (rank : 63) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
HMGA2_HUMAN
|
||||||
| NC score | 0.035164 (rank : 64) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P52926 | Gene names | HMGA2, HMGIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein HMGI-C (High mobility group AT-hook protein 2). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.034163 (rank : 65) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
REXO1_MOUSE
|
||||||
| NC score | 0.031102 (rank : 66) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TT28, Q3UMP1, Q69ZR0, Q6NSQ6, Q6PI95, Q9DA29 | Gene names | Rexo1, Kiaa1138, Tceb3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
TOP2A_MOUSE
|
||||||
| NC score | 0.029492 (rank : 67) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q01320 | Gene names | Top2a, Top-2, Top2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.027948 (rank : 68) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
ZCH11_HUMAN
|
||||||
| NC score | 0.027777 (rank : 69) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.026038 (rank : 70) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.025979 (rank : 71) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
RBM19_MOUSE
|
||||||
| NC score | 0.025907 (rank : 72) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R3C6, Q8BHR0, Q9CW63 | Gene names | Rbm19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.023327 (rank : 73) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
DDX20_MOUSE
|
||||||
| NC score | 0.021799 (rank : 74) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.020452 (rank : 75) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
BRDT_HUMAN
|
||||||
| NC score | 0.019348 (rank : 76) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
GCX1_HUMAN
|
||||||
| NC score | 0.018315 (rank : 77) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96NM4, Q5TE33, Q5TE34, Q5TE35, Q96IC9, Q9BQN5 | Gene names | GCX1, C20orf100 | |||
|
Domain Architecture |

|
|||||
| Description | Granulosa cell HMG box protein 1 (GCX-1). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.018236 (rank : 78) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
B3A3_HUMAN
|
||||||
| NC score | 0.018046 (rank : 79) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48751 | Gene names | SLC4A3, AE3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3) (Cardiac/brain band 3-like protein) (CAE3/BAE3). | |||||
|
B3A3_MOUSE
|
||||||
| NC score | 0.017868 (rank : 80) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P16283 | Gene names | Slc4a3, Ae3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3). | |||||
|
CASZ1_HUMAN
|
||||||
| NC score | 0.017642 (rank : 81) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 473 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86V15, Q5T9S1, Q8WX49, Q8WX50, Q9BT16, Q9NXC6 | Gene names | CASZ1, CST, ZNF693 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Castor homolog 1 zinc finger protein (Castor-related protein) (Zinc finger protein 693). | |||||
|
DDX20_HUMAN
|
||||||
| NC score | 0.017042 (rank : 82) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.016133 (rank : 83) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.016018 (rank : 84) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
ZN592_MOUSE
|
||||||
| NC score | 0.007934 (rank : 85) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BHZ4, Q80XM1 | Gene names | Znf592, Kiaa0211, Zfp592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 592 (Zfp-592). | |||||
|
FOG1_HUMAN
|
||||||
| NC score | 0.001943 (rank : 86) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||