Please be patient as the page loads
|
GCX1_HUMAN
|
||||||
| SwissProt Accessions | Q96NM4, Q5TE33, Q5TE34, Q5TE35, Q96IC9, Q9BQN5 | Gene names | GCX1, C20orf100 | |||
|
Domain Architecture |

|
|||||
| Description | Granulosa cell HMG box protein 1 (GCX-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GCX1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | Q96NM4, Q5TE33, Q5TE34, Q5TE35, Q96IC9, Q9BQN5 | Gene names | GCX1, C20orf100 | |||
|
Domain Architecture |

|
|||||
| Description | Granulosa cell HMG box protein 1 (GCX-1). | |||||
|
TOX_MOUSE
|
||||||
| θ value | 3.59435e-57 (rank : 2) | NC score | 0.891295 (rank : 2) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q66JW3, Q8BKH9, Q8BYQ5, Q8R4H0 | Gene names | Tox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
TOX_HUMAN
|
||||||
| θ value | 8.00737e-57 (rank : 3) | NC score | 0.888735 (rank : 3) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O94900, Q96AV5 | Gene names | TOX, KIAA0808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
CN092_MOUSE
|
||||||
| θ value | 9.52487e-34 (rank : 4) | NC score | 0.814360 (rank : 5) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
CN092_HUMAN
|
||||||
| θ value | 5.40856e-29 (rank : 5) | NC score | 0.814827 (rank : 4) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O94842 | Gene names | C14orf92, KIAA0737 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
HMGB1_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 6) | NC score | 0.509847 (rank : 7) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P09429, Q6IBE1 | Gene names | HMGB1, HMG1 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HMGB1_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 7) | NC score | 0.509989 (rank : 6) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P63158, P07155, P27109, P27428 | Gene names | Hmgb1, Hmg-1, Hmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HMG1X_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 8) | NC score | 0.497622 (rank : 8) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UGV6 | Gene names | HMG1L10 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 1-like 10 (HMG-1L10). | |||||
|
SSRP1_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 9) | NC score | 0.481897 (rank : 9) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
SSRP1_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 10) | NC score | 0.464558 (rank : 14) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
HM20B_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 11) | NC score | 0.471189 (rank : 13) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9P0W2, Q6IBP8, Q8NBD5, Q9HD21, Q9Y491, Q9Y4A2 | Gene names | HMG20B, BRAF35, HMGX2, SMARCE1R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35) (Sox-like transcriptional factor). | |||||
|
HM20B_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 12) | NC score | 0.480931 (rank : 10) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Z104, Q9NSF7, Q9NSF8 | Gene names | Hmg20b, Braf35, Hmgx2, Smarce1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 13) | NC score | 0.276671 (rank : 28) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CIC_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 14) | NC score | 0.291991 (rank : 27) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
HM20A_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 15) | NC score | 0.413161 (rank : 19) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NP66, Q53G31, Q9NSF6 | Gene names | HMG20A, HMGX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (HMG domain-containing protein HMGX1). | |||||
|
HM20A_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 16) | NC score | 0.417993 (rank : 18) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9DC33, Q3LSF9, Q6NV87, Q8BSK1, Q8C3C1, Q8CAA0, Q9CYG2 | Gene names | Hmg20a, Ibraf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (iBRAF) (Inhibitor of BRAF35) (HMG domain protein HMGX1). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 17) | NC score | 0.325788 (rank : 24) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
HMGB2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 18) | NC score | 0.475830 (rank : 11) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P26583 | Gene names | HMGB2, HMG2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 19) | NC score | 0.102006 (rank : 82) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
SP100_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 20) | NC score | 0.367070 (rank : 22) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P23497, O75450, Q13343, Q96F70, Q96T24, Q96T95, Q9NP33, Q9UE32 | Gene names | SP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein) (Lysp100b). | |||||
|
HMGB2_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 21) | NC score | 0.472798 (rank : 12) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P30681, Q3UXT1, Q9EQD5 | Gene names | Hmgb2, Hmg2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
PB1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 22) | NC score | 0.268561 (rank : 29) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 23) | NC score | 0.044505 (rank : 144) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
UBF1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 24) | NC score | 0.318017 (rank : 25) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
HMGB3_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 25) | NC score | 0.442084 (rank : 16) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O15347, O95556, Q6NS40 | Gene names | HMGB3, HMG2A, HMG4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
HMGB3_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 26) | NC score | 0.445909 (rank : 15) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O54879 | Gene names | Hmgb3, Hmg2a, Hmg4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
LCP2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 27) | NC score | 0.074693 (rank : 91) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 28) | NC score | 0.093595 (rank : 85) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TFAM_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 29) | NC score | 0.410760 (rank : 20) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q00059 | Gene names | TFAM, TCF6L2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Mitochondrial transcription factor 1) (MtTF1) (Transcription factor 6-like 2). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 30) | NC score | 0.043147 (rank : 145) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
LEF1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 31) | NC score | 0.151385 (rank : 72) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UJU2, Q9HAZ0 | Gene names | LEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1) (T cell-specific transcription factor 1-alpha) (TCF1-alpha). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 32) | NC score | 0.062705 (rank : 109) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
TULP1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 33) | NC score | 0.056936 (rank : 117) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00294, O43536 | Gene names | TULP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
LEF1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 34) | NC score | 0.151581 (rank : 71) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P27782 | Gene names | Lef1, Lef-1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 35) | NC score | 0.213080 (rank : 33) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
CBP_HUMAN
|
||||||
| θ value | 0.125558 (rank : 36) | NC score | 0.081414 (rank : 86) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CSPG3_MOUSE
|
||||||
| θ value | 0.125558 (rank : 37) | NC score | 0.025519 (rank : 159) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
HM2L1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 38) | NC score | 0.388745 (rank : 21) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UGU5, O75672, O75673, Q9UMT5 | Gene names | HMG2L1, HMGBCG | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 2-like 1 (Protein HMGBCG). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.125558 (rank : 39) | NC score | 0.062393 (rank : 110) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SMCE1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 40) | NC score | 0.332488 (rank : 23) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
SMCE1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 41) | NC score | 0.313565 (rank : 26) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.163984 (rank : 42) | NC score | 0.055643 (rank : 121) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SP5_HUMAN
|
||||||
| θ value | 0.163984 (rank : 43) | NC score | 0.009548 (rank : 188) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
TFAM_MOUSE
|
||||||
| θ value | 0.163984 (rank : 44) | NC score | 0.441323 (rank : 17) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P40630, P97894, P97906, Q543I8, Q9DBM9 | Gene names | Tfam, Hmgts | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Testis- specific high mobility group protein) (TS-HMG). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 45) | NC score | 0.068481 (rank : 99) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SOX2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 46) | NC score | 0.212648 (rank : 36) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P48431, Q14537 | Gene names | SOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 47) | NC score | 0.212705 (rank : 34) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P48432 | Gene names | Sox2, Sox-2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 48) | NC score | 0.075427 (rank : 90) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
HNF1A_MOUSE
|
||||||
| θ value | 0.279714 (rank : 49) | NC score | 0.059825 (rank : 113) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P22361 | Gene names | Tcf1, Hnf-1, Hnf-1a, Hnf1a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 50) | NC score | 0.050206 (rank : 141) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SOX7_MOUSE
|
||||||
| θ value | 0.279714 (rank : 51) | NC score | 0.227313 (rank : 30) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P40646, Q9R1T6 | Gene names | Sox7, Sox-7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7 (mSOX7). | |||||
|
SOX5_HUMAN
|
||||||
| θ value | 0.365318 (rank : 52) | NC score | 0.207339 (rank : 37) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 0.47712 (rank : 53) | NC score | 0.065096 (rank : 105) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
SOX5_MOUSE
|
||||||
| θ value | 0.47712 (rank : 54) | NC score | 0.206672 (rank : 38) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P35710, O88184, O89018 | Gene names | Sox5, Sox-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX7_HUMAN
|
||||||
| θ value | 0.47712 (rank : 55) | NC score | 0.223409 (rank : 32) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
CBP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 56) | NC score | 0.080290 (rank : 87) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
KCNQ5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 57) | NC score | 0.020173 (rank : 168) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.62314 (rank : 58) | NC score | 0.074486 (rank : 92) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SP5_MOUSE
|
||||||
| θ value | 0.62314 (rank : 59) | NC score | 0.009274 (rank : 189) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
TR95_MOUSE
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.039063 (rank : 148) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PGF3 | Gene names | Thrap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein complex 95 kDa component (Trap95) (Thyroid hormone receptor-associated protein 5) (Vitamin D3 receptor-interacting protein complex component DRIP92). | |||||
|
ANR41_HUMAN
|
||||||
| θ value | 0.813845 (rank : 61) | NC score | 0.024477 (rank : 160) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NAG6, Q8N8J8 | Gene names | ANKRD41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 41. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.813845 (rank : 62) | NC score | 0.064460 (rank : 107) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 63) | NC score | 0.039935 (rank : 147) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 64) | NC score | 0.107065 (rank : 79) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
PAX3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 65) | NC score | 0.018436 (rank : 172) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P23760, Q16448 | Gene names | PAX3, HUP2 | |||
|
Domain Architecture |
|
|||||
| Description | Paired box protein Pax-3 (HUP2). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 0.813845 (rank : 66) | NC score | 0.078360 (rank : 89) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
SOX13_HUMAN
|
||||||
| θ value | 0.813845 (rank : 67) | NC score | 0.212680 (rank : 35) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
CS007_MOUSE
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.056478 (rank : 118) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 69) | NC score | 0.038345 (rank : 149) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
PAX3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | 0.018028 (rank : 174) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P24610, Q3UFQ9, Q8BRE7, Q9CXI6 | Gene names | Pax3, Pax-3 | |||
|
Domain Architecture |
|
|||||
| Description | Paired box protein Pax-3. | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.021556 (rank : 166) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
FBLN2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.016603 (rank : 179) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
GAB2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.028593 (rank : 157) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
SOX6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.199172 (rank : 40) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P35712, Q9BXQ3, Q9BXQ4, Q9BXQ5, Q9H0I8 | Gene names | SOX6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6. | |||||
|
SOX6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.198854 (rank : 41) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P40645, Q62250, Q9QWS5 | Gene names | Sox6, Sox-6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6 (SOX-LZ). | |||||
|
SPRM1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.017180 (rank : 178) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DAC9 | Gene names | Sprm1 | |||
|
Domain Architecture |
|
|||||
| Description | Sperm 1 POU-domain transcription factor (SPRM-1). | |||||
|
FOXO4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.017916 (rank : 175) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WVH3 | Gene names | Mllt7, Afx, Afx1, Fkhr3, Foxo4 | |||
|
Domain Architecture |
|
|||||
| Description | Fork head domain transcription factor AFX1 (Afxh) (Forkhead box protein O4). | |||||
|
SOX17_MOUSE
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.189643 (rank : 50) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q61473, Q61472, Q62248 | Gene names | Sox17, Sox-17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SRY_MOUSE
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.192264 (rank : 46) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q05738 | Gene names | Sry, Tdf, Tdy | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
TTLL5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | 0.021901 (rank : 164) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
LTBP3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.009822 (rank : 187) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61810, Q8BNQ6 | Gene names | Ltbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
PMS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.177849 (rank : 54) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.040629 (rank : 146) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
SOX17_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.183259 (rank : 51) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H6I2 | Gene names | SOX17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
TULP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.049476 (rank : 142) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z273 | Gene names | Tulp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
ULK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | -0.001070 (rank : 194) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 979 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IYT8, O75119 | Gene names | ULK2, KIAA0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2). | |||||
|
ATOH1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.016386 (rank : 180) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48985 | Gene names | Atoh1, Ath1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein mATH-1) (MATH1). | |||||
|
CADH1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.003549 (rank : 193) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09803, Q61377 | Gene names | Cdh1 | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial-cadherin precursor (E-cadherin) (Uvomorulin) (Cadherin-1) (ARC-1) (CD324 antigen) [Contains: E-Cad/CTF1; E-Cad/CTF2; E- Cad/CTF3]. | |||||
|
CUL7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.019666 (rank : 170) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14999, Q5T654 | Gene names | CUL7, KIAA0076 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-7 (CUL-7). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.058360 (rank : 114) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
RB27B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.012298 (rank : 183) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00194, Q9BZB6 | Gene names | RAB27B | |||
|
Domain Architecture |
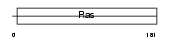
|
|||||
| Description | Ras-related protein Rab-27B (C25KG). | |||||
|
RGP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.021607 (rank : 165) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
SYT9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.008014 (rank : 190) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R0N9, Q8C280 | Gene names | Syt9, Syt5 | |||
|
Domain Architecture |
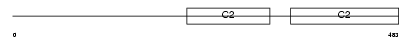
|
|||||
| Description | Synaptotagmin-9 (Synaptotagmin IX) (SytIX) (Synaptotagmin V). | |||||
|
ATX1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.057510 (rank : 116) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
II2A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.020594 (rank : 167) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15091 | Gene names | Ifi202a, Ifi202 | |||
|
Domain Architecture |
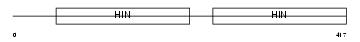
|
|||||
| Description | Interferon-activable protein 202a (Ifi-202a) (Interferon-inducible protein p202a). | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.034000 (rank : 154) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
SOX15_MOUSE
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.196354 (rank : 45) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P43267, O70204, P70418, Q62246, Q91V00, Q91V43, Q920T1, Q9JLG2 | Gene names | Sox15, Sox-15 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein. | |||||
|
CR001_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.014750 (rank : 181) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BWJ4 | Gene names | D18Ertd653e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C18orf1 homolog. | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.034200 (rank : 153) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
K0460_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.035464 (rank : 152) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
KIRR2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.005286 (rank : 191) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TSU7, Q7TQ98 | Gene names | Kirrel2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kin of IRRE-like protein 2 precursor (Kin of irregular chiasm-like protein 2). | |||||
|
NPM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.018315 (rank : 173) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06748, P08693, Q12826, Q13440, Q13441, Q14115, Q5EU94, Q5EU95, Q5EU96, Q5EU97, Q5EU98, Q5EU99, Q6V962, Q8WTW5, Q96AT6, Q96DC4, Q96EA5 | Gene names | NPM1, NPM | |||
|
Domain Architecture |

|
|||||
| Description | Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38). | |||||
|
OTP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.009849 (rank : 186) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O09113 | Gene names | Otp | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein orthopedia. | |||||
|
SOX14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.197629 (rank : 43) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O95416, Q3KPH7 | Gene names | SOX14, SOX28 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX14_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.197618 (rank : 44) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q04892, Q9JLC9, Q9JLD0, Q9QXU4 | Gene names | Sox14, Sox-14 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX30_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.198328 (rank : 42) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SOX30_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.206142 (rank : 39) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
ADAM8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.004746 (rank : 192) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q05910 | Gene names | Adam8, Ms2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 8 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 8) (Cell surface antigen MS2) (Macrophage cysteine-rich glycoprotein) (CD156 antigen). | |||||
|
ATX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.036614 (rank : 151) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
BCL6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | -0.001379 (rank : 195) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P41182 | Gene names | BCL6, BCL5, LAZ3, ZBTB27, ZNF51 | |||
|
Domain Architecture |
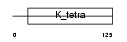
|
|||||
| Description | B-cell lymphoma 6 protein (BCL-6) (Zinc finger protein 51) (LAZ-3 protein) (BCL-5) (Zinc finger and BTB domain-containing protein 27). | |||||
|
CBL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.013335 (rank : 182) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P22681 | Gene names | CBL, CBL2, RNF55 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene) (RING finger protein 55). | |||||
|
CD248_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.022676 (rank : 163) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
DEAF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.023506 (rank : 162) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1T5 | Gene names | Deaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR). | |||||
|
NFAC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.019607 (rank : 171) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
NPAS4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.038110 (rank : 150) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
SON_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.053376 (rank : 129) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SOX15_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.189988 (rank : 49) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O60248, P35717, Q9Y6W7 | Gene names | SOX15, SOX12, SOX20, SOX26, SOX27 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein (SOX-20 protein) (SOX-12 protein). | |||||
|
SOX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.179563 (rank : 52) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O00570 | Gene names | SOX1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.179317 (rank : 53) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P53783 | Gene names | Sox1, Sox-1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SRY_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.190572 (rank : 48) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q05066 | Gene names | SRY, TDF | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
TRI66_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.032985 (rank : 156) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
ZFPL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.019761 (rank : 169) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95159, O14616, Q9UID0 | Gene names | ZFPL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein-like 1 (Zinc finger protein MCG4). | |||||
|
CDAN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.024377 (rank : 161) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IWY9, Q7Z7L5, Q969N3 | Gene names | CDAN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Codanin-1. | |||||
|
CG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.063336 (rank : 108) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
DCX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.010410 (rank : 184) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43602, O43911 | Gene names | DCX, DBCN, LISX | |||
|
Domain Architecture |
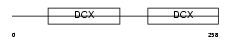
|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
DMRTC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.017288 (rank : 177) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D9R7, Q2PMY0, Q9D5U3 | Gene names | Dmrtc1, Dmrt8.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C1 (Doublesex- and mab-3-related transcription factor 8.1). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.071363 (rank : 95) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
II2B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.017425 (rank : 176) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R002 | Gene names | Ifi202b | |||
|
Domain Architecture |
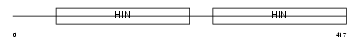
|
|||||
| Description | Interferon-activable protein 202b (Ifi-202b) (Interferon-inducible protein p202b). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.045003 (rank : 143) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
NFIC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.010152 (rank : 185) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P08651, P08652, Q14932, Q9UPJ3, Q9UPJ9, Q9UPK0, Q9UPK1 | Gene names | NFIC, NFI | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 C-type (Nuclear factor 1/C) (NF1-C) (NFI-C) (NF-I/C) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.033704 (rank : 155) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PF21B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.026649 (rank : 158) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
ALEX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.062349 (rank : 111) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.065731 (rank : 102) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.052181 (rank : 134) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.060286 (rank : 112) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BBX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.191382 (rank : 47) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WY36, Q2TAJ1, Q7L3J8, Q7LBY8, Q8NDB0, Q8WY35, Q9H0J6 | Gene names | BBX, HBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
BBX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.224983 (rank : 31) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8VBW5, Q3TZK1, Q6NXY8, Q6PEU3, Q8BQJ7, Q8C7E0, Q8CDQ0, Q8CDV1, Q8VI48, Q8VI49, Q8VI50, Q9CS94 | Gene names | Bbx, Hbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.057752 (rank : 115) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CN032_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.051368 (rank : 139) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.065673 (rank : 103) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CS016_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.055424 (rank : 123) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.053412 (rank : 128) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
EP300_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.073454 (rank : 93) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
GGN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.056143 (rank : 119) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GGN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.052867 (rank : 131) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
HBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.139574 (rank : 73) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O60381, Q8TBM1, Q8TE93, Q96AJ2 | Gene names | HBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
HBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.139332 (rank : 74) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8R316, Q3V0I4, Q8BUS3, Q8C199 | Gene names | Hbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.051405 (rank : 138) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.069829 (rank : 97) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.079301 (rank : 88) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.051608 (rank : 136) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PCQAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.053160 (rank : 130) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.051999 (rank : 135) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.055994 (rank : 120) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.054517 (rank : 126) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.068331 (rank : 100) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.065189 (rank : 104) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.070976 (rank : 96) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.054411 (rank : 127) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.051584 (rank : 137) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.051218 (rank : 140) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SOX10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.138278 (rank : 76) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
SOX10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.138838 (rank : 75) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q04888, O08518, O09141, O54856, P70416 | Gene names | Sox10, Sox-10, Sox21 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10 (SOX-21) (Transcription factor SOX-M). | |||||
|
SOX11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.160039 (rank : 66) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P35716 | Gene names | SOX11 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.162278 (rank : 61) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q7M6Y2, O35178, O89036, Q04889, Q80XF0 | Gene names | Sox11, Sox-11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.163607 (rank : 60) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O15370, Q9NUD4 | Gene names | SOX12, SOX22 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-12 protein (SOX-22 protein). | |||||
|
SOX12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.160799 (rank : 64) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q04890, P70417, Q6NXL2 | Gene names | Sox12, Sox-12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SOX-12 protein. | |||||
|
SOX18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.168000 (rank : 57) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P35713, Q9NPH8 | Gene names | SOX18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
SOX18_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.162079 (rank : 62) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P43680, Q9EQ73 | Gene names | Sox18, Sox-18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
SOX21_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.174309 (rank : 55) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y651, P35715, Q15504 | Gene names | SOX21, SOX25, SOXA | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-21 (SOX-A). | |||||
|
SOX21_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.174263 (rank : 56) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q811W0, Q8VH35 | Gene names | Sox21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-21. | |||||
|
SOX3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.164248 (rank : 59) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P41225, P35714, Q9NP49 | Gene names | SOX3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SOX3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.166538 (rank : 58) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P53784 | Gene names | Sox3, Sox-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SOX4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.161481 (rank : 63) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q06945 | Gene names | SOX4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.160756 (rank : 65) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q06831 | Gene names | Sox4, Sox-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.156778 (rank : 67) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P57073, Q9NZW2 | Gene names | SOX8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.155994 (rank : 68) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q04886 | Gene names | Sox8, Sox-8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.152700 (rank : 69) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.152193 (rank : 70) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q04887, Q8C7L2, Q91ZK2, Q99KQ0 | Gene names | Sox9, Sox-9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.068293 (rank : 101) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.055309 (rank : 124) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.073413 (rank : 94) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TCF7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.120425 (rank : 77) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P36402, Q9UKI4 | Gene names | TCF7, TCF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
TCF7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.116869 (rank : 78) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
TF7L1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.101533 (rank : 83) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9HCS4, Q53R97, Q6PD70, Q9NP00 | Gene names | TCF7L1, TCF3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF- 3). | |||||
|
TF7L1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.100548 (rank : 84) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z1J1, O70450, O70573 | Gene names | Tcf7l1, Tcf3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF-3) (mTCF-3). | |||||
|
TF7L2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.103421 (rank : 81) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NQB0, O00185, Q9NQB1, Q9NQB2, Q9NQB3, Q9NQB4, Q9NQB5, Q9NQB6, Q9NQB7, Q9ULC2 | Gene names | TCF7L2, TCF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (hTCF-4). | |||||
|
TF7L2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.104713 (rank : 80) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q924A0, O70574, Q91XP2, Q91XP3, Q91XP4, Q924A1, Q9Z0V3, Q9Z0V4 | Gene names | Tcf7l2, Tcf4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (mTCF-4). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.069425 (rank : 98) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.065060 (rank : 106) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.055273 (rank : 125) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.052390 (rank : 133) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.055553 (rank : 122) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ZN207_HUMAN
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.052546 (rank : 132) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
GCX1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | Q96NM4, Q5TE33, Q5TE34, Q5TE35, Q96IC9, Q9BQN5 | Gene names | GCX1, C20orf100 | |||
|
Domain Architecture |

|
|||||
| Description | Granulosa cell HMG box protein 1 (GCX-1). | |||||
|
TOX_MOUSE
|
||||||
| NC score | 0.891295 (rank : 2) | θ value | 3.59435e-57 (rank : 2) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q66JW3, Q8BKH9, Q8BYQ5, Q8R4H0 | Gene names | Tox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
TOX_HUMAN
|
||||||
| NC score | 0.888735 (rank : 3) | θ value | 8.00737e-57 (rank : 3) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O94900, Q96AV5 | Gene names | TOX, KIAA0808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
CN092_HUMAN
|
||||||
| NC score | 0.814827 (rank : 4) | θ value | 5.40856e-29 (rank : 5) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O94842 | Gene names | C14orf92, KIAA0737 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
CN092_MOUSE
|
||||||
| NC score | 0.814360 (rank : 5) | θ value | 9.52487e-34 (rank : 4) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
HMGB1_MOUSE
|
||||||
| NC score | 0.509989 (rank : 6) | θ value | 1.38499e-08 (rank : 7) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P63158, P07155, P27109, P27428 | Gene names | Hmgb1, Hmg-1, Hmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HMGB1_HUMAN
|
||||||
| NC score | 0.509847 (rank : 7) | θ value | 1.38499e-08 (rank : 6) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P09429, Q6IBE1 | Gene names | HMGB1, HMG1 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HMG1X_HUMAN
|
||||||
| NC score | 0.497622 (rank : 8) | θ value | 3.41135e-07 (rank : 8) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UGV6 | Gene names | HMG1L10 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 1-like 10 (HMG-1L10). | |||||
|
SSRP1_HUMAN
|
||||||
| NC score | 0.481897 (rank : 9) | θ value | 2.88788e-06 (rank : 9) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
HM20B_MOUSE
|
||||||
| NC score | 0.480931 (rank : 10) | θ value | 0.000461057 (rank : 12) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Z104, Q9NSF7, Q9NSF8 | Gene names | Hmg20b, Braf35, Hmgx2, Smarce1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35). | |||||
|
HMGB2_HUMAN
|
||||||
| NC score | 0.475830 (rank : 11) | θ value | 0.00390308 (rank : 18) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P26583 | Gene names | HMGB2, HMG2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
HMGB2_MOUSE
|
||||||
| NC score | 0.472798 (rank : 12) | θ value | 0.00665767 (rank : 21) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P30681, Q3UXT1, Q9EQD5 | Gene names | Hmgb2, Hmg2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
HM20B_HUMAN
|
||||||
| NC score | 0.471189 (rank : 13) | θ value | 0.000461057 (rank : 11) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9P0W2, Q6IBP8, Q8NBD5, Q9HD21, Q9Y491, Q9Y4A2 | Gene names | HMG20B, BRAF35, HMGX2, SMARCE1R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35) (Sox-like transcriptional factor). | |||||
|
SSRP1_MOUSE
|
||||||
| NC score | 0.464558 (rank : 14) | θ value | 9.29e-05 (rank : 10) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
HMGB3_MOUSE
|
||||||
| NC score | 0.445909 (rank : 15) | θ value | 0.0193708 (rank : 26) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O54879 | Gene names | Hmgb3, Hmg2a, Hmg4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
HMGB3_HUMAN
|
||||||
| NC score | 0.442084 (rank : 16) | θ value | 0.0193708 (rank : 25) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O15347, O95556, Q6NS40 | Gene names | HMGB3, HMG2A, HMG4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
TFAM_MOUSE
|
||||||
| NC score | 0.441323 (rank : 17) | θ value | 0.163984 (rank : 44) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P40630, P97894, P97906, Q543I8, Q9DBM9 | Gene names | Tfam, Hmgts | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Testis- specific high mobility group protein) (TS-HMG). | |||||
|
HM20A_MOUSE
|
||||||
| NC score | 0.417993 (rank : 18) | θ value | 0.00175202 (rank : 16) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9DC33, Q3LSF9, Q6NV87, Q8BSK1, Q8C3C1, Q8CAA0, Q9CYG2 | Gene names | Hmg20a, Ibraf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (iBRAF) (Inhibitor of BRAF35) (HMG domain protein HMGX1). | |||||
|
HM20A_HUMAN
|
||||||
| NC score | 0.413161 (rank : 19) | θ value | 0.00175202 (rank : 15) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NP66, Q53G31, Q9NSF6 | Gene names | HMG20A, HMGX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (HMG domain-containing protein HMGX1). | |||||
|
TFAM_HUMAN
|
||||||
| NC score | 0.410760 (rank : 20) | θ value | 0.0431538 (rank : 29) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q00059 | Gene names | TFAM, TCF6L2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Mitochondrial transcription factor 1) (MtTF1) (Transcription factor 6-like 2). | |||||
|
HM2L1_HUMAN
|
||||||
| NC score | 0.388745 (rank : 21) | θ value | 0.125558 (rank : 38) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UGU5, O75672, O75673, Q9UMT5 | Gene names | HMG2L1, HMGBCG | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 2-like 1 (Protein HMGBCG). | |||||
|
SP100_HUMAN
|
||||||
| NC score | 0.367070 (rank : 22) | θ value | 0.00509761 (rank : 20) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P23497, O75450, Q13343, Q96F70, Q96T24, Q96T95, Q9NP33, Q9UE32 | Gene names | SP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein) (Lysp100b). | |||||
|
SMCE1_HUMAN
|
||||||
| NC score | 0.332488 (rank : 23) | θ value | 0.125558 (rank : 40) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.325788 (rank : 24) | θ value | 0.00175202 (rank : 17) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
UBF1_MOUSE
|
||||||
| NC score | 0.318017 (rank : 25) | θ value | 0.00869519 (rank : 24) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
SMCE1_MOUSE
|
||||||
| NC score | 0.313565 (rank : 26) | θ value | 0.125558 (rank : 41) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.291991 (rank : 27) | θ value | 0.00175202 (rank : 14) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.276671 (rank : 28) | θ value | 0.00175202 (rank : 13) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
PB1_HUMAN
|
||||||
| NC score | 0.268561 (rank : 29) | θ value | 0.00665767 (rank : 22) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
SOX7_MOUSE
|
||||||
| NC score | 0.227313 (rank : 30) | θ value | 0.279714 (rank : 51) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P40646, Q9R1T6 | Gene names | Sox7, Sox-7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7 (mSOX7). | |||||
|
BBX_MOUSE
|
||||||
| NC score | 0.224983 (rank : 31) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8VBW5, Q3TZK1, Q6NXY8, Q6PEU3, Q8BQJ7, Q8C7E0, Q8CDQ0, Q8CDV1, Q8VI48, Q8VI49, Q8VI50, Q9CS94 | Gene names | Bbx, Hbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
SOX7_HUMAN
|
||||||
| NC score | 0.223409 (rank : 32) | θ value | 0.47712 (rank : 55) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.213080 (rank : 33) | θ value | 0.0961366 (rank : 35) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SOX2_MOUSE
|
||||||
| NC score | 0.212705 (rank : 34) | θ value | 0.21417 (rank : 47) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P48432 | Gene names | Sox2, Sox-2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX13_HUMAN
|
||||||
| NC score | 0.212680 (rank : 35) | θ value | 0.813845 (rank : 67) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
SOX2_HUMAN
|
||||||
| NC score | 0.212648 (rank : 36) | θ value | 0.21417 (rank : 46) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P48431, Q14537 | Gene names | SOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX5_HUMAN
|
||||||
| NC score | 0.207339 (rank : 37) | θ value | 0.365318 (rank : 52) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX5_MOUSE
|
||||||
| NC score | 0.206672 (rank : 38) | θ value | 0.47712 (rank : 54) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P35710, O88184, O89018 | Gene names | Sox5, Sox-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX30_MOUSE
|
||||||
| NC score | 0.206142 (rank : 39) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SOX6_HUMAN
|
||||||
| NC score | 0.199172 (rank : 40) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P35712, Q9BXQ3, Q9BXQ4, Q9BXQ5, Q9H0I8 | Gene names | SOX6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6. | |||||
|
SOX6_MOUSE
|
||||||
| NC score | 0.198854 (rank : 41) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P40645, Q62250, Q9QWS5 | Gene names | Sox6, Sox-6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6 (SOX-LZ). | |||||
|
SOX30_HUMAN
|
||||||
| NC score | 0.198328 (rank : 42) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SOX14_HUMAN
|
||||||
| NC score | 0.197629 (rank : 43) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O95416, Q3KPH7 | Gene names | SOX14, SOX28 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX14_MOUSE
|
||||||
| NC score | 0.197618 (rank : 44) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q04892, Q9JLC9, Q9JLD0, Q9QXU4 | Gene names | Sox14, Sox-14 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX15_MOUSE
|
||||||
| NC score | 0.196354 (rank : 45) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P43267, O70204, P70418, Q62246, Q91V00, Q91V43, Q920T1, Q9JLG2 | Gene names | Sox15, Sox-15 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein. | |||||
|
SRY_MOUSE
|
||||||
| NC score | 0.192264 (rank : 46) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q05738 | Gene names | Sry, Tdf, Tdy | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
BBX_HUMAN
|
||||||
| NC score | 0.191382 (rank : 47) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WY36, Q2TAJ1, Q7L3J8, Q7LBY8, Q8NDB0, Q8WY35, Q9H0J6 | Gene names | BBX, HBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
SRY_HUMAN
|
||||||
| NC score | 0.190572 (rank : 48) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q05066 | Gene names | SRY, TDF | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
SOX15_HUMAN
|
||||||
| NC score | 0.189988 (rank : 49) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O60248, P35717, Q9Y6W7 | Gene names | SOX15, SOX12, SOX20, SOX26, SOX27 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein (SOX-20 protein) (SOX-12 protein). | |||||
|
SOX17_MOUSE
|
||||||
| NC score | 0.189643 (rank : 50) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q61473, Q61472, Q62248 | Gene names | Sox17, Sox-17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX17_HUMAN
|
||||||
| NC score | 0.183259 (rank : 51) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H6I2 | Gene names | SOX17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX1_HUMAN
|
||||||
| NC score | 0.179563 (rank : 52) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O00570 | Gene names | SOX1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX1_MOUSE
|
||||||
| NC score | 0.179317 (rank : 53) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P53783 | Gene names | Sox1, Sox-1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
PMS1_HUMAN
|
||||||
| NC score | 0.177849 (rank : 54) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
SOX21_HUMAN
|
||||||
| NC score | 0.174309 (rank : 55) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y651, P35715, Q15504 | Gene names | SOX21, SOX25, SOXA | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-21 (SOX-A). | |||||
|
SOX21_MOUSE
|
||||||
| NC score | 0.174263 (rank : 56) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q811W0, Q8VH35 | Gene names | Sox21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-21. | |||||
|
SOX18_HUMAN
|
||||||
| NC score | 0.168000 (rank : 57) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P35713, Q9NPH8 | Gene names | SOX18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
SOX3_MOUSE
|
||||||
| NC score | 0.166538 (rank : 58) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P53784 | Gene names | Sox3, Sox-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SOX3_HUMAN
|
||||||
| NC score | 0.164248 (rank : 59) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P41225, P35714, Q9NP49 | Gene names | SOX3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SOX12_HUMAN
|
||||||
| NC score | 0.163607 (rank : 60) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O15370, Q9NUD4 | Gene names | SOX12, SOX22 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-12 protein (SOX-22 protein). | |||||
|
SOX11_MOUSE
|
||||||
| NC score | 0.162278 (rank : 61) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q7M6Y2, O35178, O89036, Q04889, Q80XF0 | Gene names | Sox11, Sox-11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX18_MOUSE
|
||||||
| NC score | 0.162079 (rank : 62) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P43680, Q9EQ73 | Gene names | Sox18, Sox-18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
SOX4_HUMAN
|
||||||
| NC score | 0.161481 (rank : 63) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q06945 | Gene names | SOX4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX12_MOUSE
|
||||||
| NC score | 0.160799 (rank : 64) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q04890, P70417, Q6NXL2 | Gene names | Sox12, Sox-12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SOX-12 protein. | |||||
|
SOX4_MOUSE
|
||||||
| NC score | 0.160756 (rank : 65) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q06831 | Gene names | Sox4, Sox-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX11_HUMAN
|
||||||
| NC score | 0.160039 (rank : 66) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P35716 | Gene names | SOX11 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX8_HUMAN
|
||||||
| NC score | 0.156778 (rank : 67) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P57073, Q9NZW2 | Gene names | SOX8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX8_MOUSE
|
||||||
| NC score | 0.155994 (rank : 68) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q04886 | Gene names | Sox8, Sox-8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX9_HUMAN
|
||||||
| NC score | 0.152700 (rank : 69) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX9_MOUSE
|
||||||
| NC score | 0.152193 (rank : 70) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q04887, Q8C7L2, Q91ZK2, Q99KQ0 | Gene names | Sox9, Sox-9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
LEF1_MOUSE
|
||||||
| NC score | 0.151581 (rank : 71) | θ value | 0.0961366 (rank : 34) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P27782 | Gene names | Lef1, Lef-1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1). | |||||
|
LEF1_HUMAN
|
||||||
| NC score | 0.151385 (rank : 72) | θ value | 0.0736092 (rank : 31) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UJU2, Q9HAZ0 | Gene names | LEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1) (T cell-specific transcription factor 1-alpha) (TCF1-alpha). | |||||
|
HBP1_HUMAN
|
||||||
| NC score | 0.139574 (rank : 73) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O60381, Q8TBM1, Q8TE93, Q96AJ2 | Gene names | HBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
HBP1_MOUSE
|
||||||
| NC score | 0.139332 (rank : 74) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8R316, Q3V0I4, Q8BUS3, Q8C199 | Gene names | Hbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
SOX10_MOUSE
|
||||||
| NC score | 0.138838 (rank : 75) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q04888, O08518, O09141, O54856, P70416 | Gene names | Sox10, Sox-10, Sox21 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10 (SOX-21) (Transcription factor SOX-M). | |||||
|
SOX10_HUMAN
|
||||||
| NC score | 0.138278 (rank : 76) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
TCF7_HUMAN
|
||||||
| NC score | 0.120425 (rank : 77) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P36402, Q9UKI4 | Gene names | TCF7, TCF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
TCF7_MOUSE
|
||||||
| NC score | 0.116869 (rank : 78) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.107065 (rank : 79) | θ value | 0.813845 (rank : 64) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
TF7L2_MOUSE
|
||||||
| NC score | 0.104713 (rank : 80) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q924A0, O70574, Q91XP2, Q91XP3, Q91XP4, Q924A1, Q9Z0V3, Q9Z0V4 | Gene names | Tcf7l2, Tcf4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (mTCF-4). | |||||
|
TF7L2_HUMAN
|
||||||
| NC score | 0.103421 (rank : 81) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NQB0, O00185, Q9NQB1, Q9NQB2, Q9NQB3, Q9NQB4, Q9NQB5, Q9NQB6, Q9NQB7, Q9ULC2 | Gene names | TCF7L2, TCF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (hTCF-4). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.102006 (rank : 82) | θ value | 0.00390308 (rank : 19) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
TF7L1_HUMAN
|
||||||
| NC score | 0.101533 (rank : 83) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9HCS4, Q53R97, Q6PD70, Q9NP00 | Gene names | TCF7L1, TCF3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF- 3). | |||||
|
TF7L1_MOUSE
|
||||||
| NC score | 0.100548 (rank : 84) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z1J1, O70450, O70573 | Gene names | Tcf7l1, Tcf3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF-3) (mTCF-3). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.093595 (rank : 85) | θ value | 0.0431538 (rank : 28) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.081414 (rank : 86) | θ value | 0.125558 (rank : 36) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.080290 (rank : 87) | θ value | 0.62314 (rank : 56) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.079301 (rank : 88) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.078360 (rank : 89) | θ value | 0.813845 (rank : 66) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.075427 (rank : 90) | θ value | 0.279714 (rank : 48) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
LCP2_HUMAN
|
||||||
| NC score | 0.074693 (rank : 91) | θ value | 0.0193708 (rank : 27) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.074486 (rank : 92) | θ value | 0.62314 (rank : 58) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.073454 (rank : 93) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.073413 (rank : 94) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.071363 (rank : 95) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.070976 (rank : 96) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.069829 (rank : 97) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.069425 (rank : 98) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.068481 (rank : 99) | θ value | 0.21417 (rank : 45) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.068331 (rank : 100) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.068293 (rank : 101) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.065731 (rank : 102) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.065673 (rank : 103) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.065189 (rank : 104) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.065096 (rank : 105) | θ value | 0.47712 (rank : 53) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.065060 (rank : 106) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.064460 (rank : 107) | θ value | 0.813845 (rank : 62) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CG1_HUMAN
|
||||||
| NC score | 0.063336 (rank : 108) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.062705 (rank : 109) | θ value | 0.0736092 (rank : 32) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.062393 (rank : 110) | θ value | 0.125558 (rank : 39) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.062349 (rank : 111) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.060286 (rank : 112) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
HNF1A_MOUSE
|
||||||
| NC score | 0.059825 (rank : 113) | θ value | 0.279714 (rank : 49) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P22361 | Gene names | Tcf1, Hnf-1, Hnf-1a, Hnf1a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.058360 (rank : 114) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.057752 (rank : 115) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
ATX1_MOUSE
|
||||||
| NC score | 0.057510 (rank : 116) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
TULP1_HUMAN
|
||||||
| NC score | 0.056936 (rank : 117) | θ value | 0.0736092 (rank : 33) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00294, O43536 | Gene names | TULP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.056478 (rank : 118) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
GGN_HUMAN
|
||||||
| NC score | 0.056143 (rank : 119) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.055994 (rank : 120) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.055643 (rank : 121) | θ value | 0.163984 (rank : 42) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.055553 (rank : 122) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.055424 (rank : 123) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.055309 (rank : 124) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.055273 (rank : 125) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.054517 (rank : 126) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.054411 (rank : 127) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.053412 (rank : 128) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.053376 (rank : 129) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
PCQAP_HUMAN
|
||||||
| NC score | 0.053160 (rank : 130) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.052867 (rank : 131) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.052546 (rank : 132) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.052390 (rank : 133) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.052181 (rank : 134) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.051999 (rank : 135) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.051608 (rank : 136) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.051584 (rank : 137) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.051405 (rank : 138) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.051368 (rank : 139) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.051218 (rank : 140) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.050206 (rank : 141) | θ value | 0.279714 (rank : 50) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
TULP1_MOUSE
|
||||||
| NC score | 0.049476 (rank : 142) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z273 | Gene names | Tulp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.045003 (rank : 143) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.044505 (rank : 144) | θ value | 0.00869519 (rank : 23) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.043147 (rank : 145) | θ value | 0.0431538 (rank : 30) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.040629 (rank : 146) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.039935 (rank : 147) | θ value | 0.813845 (rank : 63) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
TR95_MOUSE
|
||||||
| NC score | 0.039063 (rank : 148) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PGF3 | Gene names | Thrap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein complex 95 kDa component (Trap95) (Thyroid hormone receptor-associated protein 5) (Vitamin D3 receptor-interacting protein complex component DRIP92). | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.038345 (rank : 149) | θ value | 1.06291 (rank : 69) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
NPAS4_MOUSE
|
||||||
| NC score | 0.038110 (rank : 150) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
ATX2_HUMAN
|
||||||
| NC score | 0.036614 (rank : 151) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.035464 (rank : 152) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.034200 (rank : 153) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.034000 (rank : 154) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.033704 (rank : 155) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.032985 (rank : 156) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
GAB2_MOUSE
|
||||||
| NC score | 0.028593 (rank : 157) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.026649 (rank : 158) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
CSPG3_MOUSE
|
||||||
| NC score | 0.025519 (rank : 159) | θ value | 0.125558 (rank : 37) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
ANR41_HUMAN
|
||||||
| NC score | 0.024477 (rank : 160) | θ value | 0.813845 (rank : 61) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NAG6, Q8N8J8 | Gene names | ANKRD41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 41. | |||||
|
CDAN1_HUMAN
|
||||||
| NC score | 0.024377 (rank : 161) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IWY9, Q7Z7L5, Q969N3 | Gene names | CDAN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Codanin-1. | |||||
|
DEAF1_MOUSE
|
||||||
| NC score | 0.023506 (rank : 162) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1T5 | Gene names | Deaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR). | |||||
|
CD248_MOUSE
|
||||||
| NC score | 0.022676 (rank : 163) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
TTLL5_MOUSE
|
||||||
| NC score | 0.021901 (rank : 164) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
RGP1_MOUSE
|
||||||
| NC score | 0.021607 (rank : 165) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.021556 (rank : 166) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
II2A_MOUSE
|
||||||
| NC score | 0.020594 (rank : 167) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15091 | Gene names | Ifi202a, Ifi202 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 202a (Ifi-202a) (Interferon-inducible protein p202a). | |||||
|
KCNQ5_HUMAN
|
||||||
| NC score | 0.020173 (rank : 168) | θ value | 0.62314 (rank : 57) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
ZFPL1_HUMAN
|
||||||
| NC score | 0.019761 (rank : 169) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95159, O14616, Q9UID0 | Gene names | ZFPL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein-like 1 (Zinc finger protein MCG4). | |||||
|
CUL7_HUMAN
|
||||||
| NC score | 0.019666 (rank : 170) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14999, Q5T654 | Gene names | CUL7, KIAA0076 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-7 (CUL-7). | |||||
|
NFAC2_HUMAN
|
||||||
| NC score | 0.019607 (rank : 171) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
PAX3_HUMAN
|
||||||
| NC score | 0.018436 (rank : 172) | θ value | 0.813845 (rank : 65) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P23760, Q16448 | Gene names | PAX3, HUP2 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-3 (HUP2). | |||||
|
NPM_HUMAN
|
||||||
| NC score | 0.018315 (rank : 173) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06748, P08693, Q12826, Q13440, Q13441, Q14115, Q5EU94, Q5EU95, Q5EU96, Q5EU97, Q5EU98, Q5EU99, Q6V962, Q8WTW5, Q96AT6, Q96DC4, Q96EA5 | Gene names | NPM1, NPM | |||
|
Domain Architecture |

|
|||||
| Description | Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38). | |||||
|
PAX3_MOUSE
|
||||||
| NC score | 0.018028 (rank : 174) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P24610, Q3UFQ9, Q8BRE7, Q9CXI6 | Gene names | Pax3, Pax-3 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-3. | |||||
|
FOXO4_MOUSE
|
||||||
| NC score | 0.017916 (rank : 175) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WVH3 | Gene names | Mllt7, Afx, Afx1, Fkhr3, Foxo4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Afxh) (Forkhead box protein O4). | |||||
|
II2B_MOUSE
|
||||||
| NC score | 0.017425 (rank : 176) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R002 | Gene names | Ifi202b | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 202b (Ifi-202b) (Interferon-inducible protein p202b). | |||||
|
DMRTC_MOUSE
|
||||||
| NC score | 0.017288 (rank : 177) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D9R7, Q2PMY0, Q9D5U3 | Gene names | Dmrtc1, Dmrt8.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C1 (Doublesex- and mab-3-related transcription factor 8.1). | |||||
|
SPRM1_MOUSE
|
||||||
| NC score | 0.017180 (rank : 178) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DAC9 | Gene names | Sprm1 | |||
|
Domain Architecture |

|
|||||
| Description | Sperm 1 POU-domain transcription factor (SPRM-1). | |||||
|
FBLN2_MOUSE
|
||||||
| NC score | 0.016603 (rank : 179) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
ATOH1_MOUSE
|
||||||
| NC score | 0.016386 (rank : 180) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48985 | Gene names | Atoh1, Ath1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein mATH-1) (MATH1). | |||||
|
CR001_MOUSE
|
||||||
| NC score | 0.014750 (rank : 181) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BWJ4 | Gene names | D18Ertd653e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C18orf1 homolog. | |||||
|
CBL_HUMAN
|
||||||
| NC score | 0.013335 (rank : 182) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P22681 | Gene names | CBL, CBL2, RNF55 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene) (RING finger protein 55). | |||||
|
RB27B_HUMAN
|
||||||
| NC score | 0.012298 (rank : 183) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00194, Q9BZB6 | Gene names | RAB27B | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-27B (C25KG). | |||||
|
DCX_HUMAN
|
||||||
| NC score | 0.010410 (rank : 184) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43602, O43911 | Gene names | DCX, DBCN, LISX | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
NFIC_HUMAN
|
||||||
| NC score | 0.010152 (rank : 185) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P08651, P08652, Q14932, Q9UPJ3, Q9UPJ9, Q9UPK0, Q9UPK1 | Gene names | NFIC, NFI | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 C-type (Nuclear factor 1/C) (NF1-C) (NFI-C) (NF-I/C) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
OTP_MOUSE
|
||||||
| NC score | 0.009849 (rank : 186) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O09113 | Gene names | Otp | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein orthopedia. | |||||
|
LTBP3_MOUSE
|
||||||
| NC score | 0.009822 (rank : 187) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61810, Q8BNQ6 | Gene names | Ltbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
SP5_HUMAN
|
||||||
| NC score | 0.009548 (rank : 188) | θ value | 0.163984 (rank : 43) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
SP5_MOUSE
|
||||||
| NC score | 0.009274 (rank : 189) | θ value | 0.62314 (rank : 59) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
SYT9_MOUSE
|
||||||
| NC score | 0.008014 (rank : 190) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R0N9, Q8C280 | Gene names | Syt9, Syt5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-9 (Synaptotagmin IX) (SytIX) (Synaptotagmin V). | |||||
|
KIRR2_MOUSE
|
||||||
| NC score | 0.005286 (rank : 191) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TSU7, Q7TQ98 | Gene names | Kirrel2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kin of IRRE-like protein 2 precursor (Kin of irregular chiasm-like protein 2). | |||||
|
ADAM8_MOUSE
|
||||||
| NC score | 0.004746 (rank : 192) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q05910 | Gene names | Adam8, Ms2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 8 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 8) (Cell surface antigen MS2) (Macrophage cysteine-rich glycoprotein) (CD156 antigen). | |||||
|
CADH1_MOUSE
|
||||||
| NC score | 0.003549 (rank : 193) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09803, Q61377 | Gene names | Cdh1 | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial-cadherin precursor (E-cadherin) (Uvomorulin) (Cadherin-1) (ARC-1) (CD324 antigen) [Contains: E-Cad/CTF1; E-Cad/CTF2; E- Cad/CTF3]. | |||||
|
ULK2_HUMAN
|
||||||
| NC score | -0.001070 (rank : 194) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 979 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IYT8, O75119 | Gene names | ULK2, KIAA0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2). | |||||
|
BCL6_HUMAN
|
||||||
| NC score | -0.001379 (rank : 195) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P41182 | Gene names | BCL6, BCL5, LAZ3, ZBTB27, ZNF51 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell lymphoma 6 protein (BCL-6) (Zinc finger protein 51) (LAZ-3 protein) (BCL-5) (Zinc finger and BTB domain-containing protein 27). | |||||