Please be patient as the page loads
|
SOX7_HUMAN
|
||||||
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SOX7_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
SOX7_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.993655 (rank : 2) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P40646, Q9R1T6 | Gene names | Sox7, Sox-7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7 (mSOX7). | |||||
|
SOX17_HUMAN
|
||||||
| θ value | 3.47335e-60 (rank : 3) | NC score | 0.967026 (rank : 5) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9H6I2 | Gene names | SOX17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX17_MOUSE
|
||||||
| θ value | 1.23536e-57 (rank : 4) | NC score | 0.954851 (rank : 6) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q61473, Q61472, Q62248 | Gene names | Sox17, Sox-17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX18_MOUSE
|
||||||
| θ value | 8.00737e-57 (rank : 5) | NC score | 0.970597 (rank : 3) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P43680, Q9EQ73 | Gene names | Sox18, Sox-18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
SOX18_HUMAN
|
||||||
| θ value | 1.78386e-56 (rank : 6) | NC score | 0.968104 (rank : 4) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P35713, Q9NPH8 | Gene names | SOX18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
SOX1_MOUSE
|
||||||
| θ value | 1.57365e-28 (rank : 7) | NC score | 0.927013 (rank : 21) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P53783 | Gene names | Sox1, Sox-1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX1_HUMAN
|
||||||
| θ value | 3.87602e-27 (rank : 8) | NC score | 0.927296 (rank : 20) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | O00570 | Gene names | SOX1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX9_HUMAN
|
||||||
| θ value | 6.61148e-27 (rank : 9) | NC score | 0.892600 (rank : 32) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX9_MOUSE
|
||||||
| θ value | 6.61148e-27 (rank : 10) | NC score | 0.893738 (rank : 31) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q04887, Q8C7L2, Q91ZK2, Q99KQ0 | Gene names | Sox9, Sox-9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX11_HUMAN
|
||||||
| θ value | 8.63488e-27 (rank : 11) | NC score | 0.936859 (rank : 10) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P35716 | Gene names | SOX11 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX10_HUMAN
|
||||||
| θ value | 2.51237e-26 (rank : 12) | NC score | 0.895674 (rank : 30) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
SOX10_MOUSE
|
||||||
| θ value | 2.51237e-26 (rank : 13) | NC score | 0.896002 (rank : 29) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q04888, O08518, O09141, O54856, P70416 | Gene names | Sox10, Sox-10, Sox21 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10 (SOX-21) (Transcription factor SOX-M). | |||||
|
SOX11_MOUSE
|
||||||
| θ value | 2.51237e-26 (rank : 14) | NC score | 0.933039 (rank : 12) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q7M6Y2, O35178, O89036, Q04889, Q80XF0 | Gene names | Sox11, Sox-11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX2_MOUSE
|
||||||
| θ value | 2.51237e-26 (rank : 15) | NC score | 0.924085 (rank : 23) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P48432 | Gene names | Sox2, Sox-2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX3_MOUSE
|
||||||
| θ value | 2.51237e-26 (rank : 16) | NC score | 0.925439 (rank : 22) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P53784 | Gene names | Sox3, Sox-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SOX8_HUMAN
|
||||||
| θ value | 3.28125e-26 (rank : 17) | NC score | 0.915241 (rank : 27) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P57073, Q9NZW2 | Gene names | SOX8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX3_HUMAN
|
||||||
| θ value | 4.28545e-26 (rank : 18) | NC score | 0.921313 (rank : 25) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P41225, P35714, Q9NP49 | Gene names | SOX3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SOX14_MOUSE
|
||||||
| θ value | 5.59698e-26 (rank : 19) | NC score | 0.927618 (rank : 18) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q04892, Q9JLC9, Q9JLD0, Q9QXU4 | Gene names | Sox14, Sox-14 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX2_HUMAN
|
||||||
| θ value | 5.59698e-26 (rank : 20) | NC score | 0.923775 (rank : 24) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P48431, Q14537 | Gene names | SOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX14_HUMAN
|
||||||
| θ value | 7.30988e-26 (rank : 21) | NC score | 0.927462 (rank : 19) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | O95416, Q3KPH7 | Gene names | SOX14, SOX28 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX15_HUMAN
|
||||||
| θ value | 7.30988e-26 (rank : 22) | NC score | 0.930373 (rank : 16) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O60248, P35717, Q9Y6W7 | Gene names | SOX15, SOX12, SOX20, SOX26, SOX27 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein (SOX-20 protein) (SOX-12 protein). | |||||
|
SOX8_MOUSE
|
||||||
| θ value | 7.30988e-26 (rank : 23) | NC score | 0.913719 (rank : 28) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q04886 | Gene names | Sox8, Sox-8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX12_MOUSE
|
||||||
| θ value | 1.62847e-25 (rank : 24) | NC score | 0.945892 (rank : 7) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q04890, P70417, Q6NXL2 | Gene names | Sox12, Sox-12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SOX-12 protein. | |||||
|
SOX12_HUMAN
|
||||||
| θ value | 2.77775e-25 (rank : 25) | NC score | 0.943009 (rank : 8) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O15370, Q9NUD4 | Gene names | SOX12, SOX22 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-12 protein (SOX-22 protein). | |||||
|
SOX4_MOUSE
|
||||||
| θ value | 3.62785e-25 (rank : 26) | NC score | 0.928558 (rank : 17) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q06831 | Gene names | Sox4, Sox-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX4_HUMAN
|
||||||
| θ value | 4.73814e-25 (rank : 27) | NC score | 0.931224 (rank : 15) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q06945 | Gene names | SOX4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX21_HUMAN
|
||||||
| θ value | 1.05554e-24 (rank : 28) | NC score | 0.932730 (rank : 14) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9Y651, P35715, Q15504 | Gene names | SOX21, SOX25, SOXA | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-21 (SOX-A). | |||||
|
SOX21_MOUSE
|
||||||
| θ value | 1.05554e-24 (rank : 29) | NC score | 0.932808 (rank : 13) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q811W0, Q8VH35 | Gene names | Sox21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-21. | |||||
|
SOX15_MOUSE
|
||||||
| θ value | 3.07116e-24 (rank : 30) | NC score | 0.934405 (rank : 11) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P43267, O70204, P70418, Q62246, Q91V00, Q91V43, Q920T1, Q9JLG2 | Gene names | Sox15, Sox-15 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein. | |||||
|
SOX5_HUMAN
|
||||||
| θ value | 6.84181e-24 (rank : 31) | NC score | 0.846493 (rank : 34) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX5_MOUSE
|
||||||
| θ value | 6.84181e-24 (rank : 32) | NC score | 0.849123 (rank : 33) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P35710, O88184, O89018 | Gene names | Sox5, Sox-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SRY_MOUSE
|
||||||
| θ value | 2.59989e-23 (rank : 33) | NC score | 0.919150 (rank : 26) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q05738 | Gene names | Sry, Tdf, Tdy | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
SRY_HUMAN
|
||||||
| θ value | 5.79196e-23 (rank : 34) | NC score | 0.940027 (rank : 9) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q05066 | Gene names | SRY, TDF | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
SOX13_HUMAN
|
||||||
| θ value | 1.29031e-22 (rank : 35) | NC score | 0.821028 (rank : 38) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
SOX6_HUMAN
|
||||||
| θ value | 1.29031e-22 (rank : 36) | NC score | 0.827922 (rank : 35) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P35712, Q9BXQ3, Q9BXQ4, Q9BXQ5, Q9H0I8 | Gene names | SOX6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6. | |||||
|
SOX6_MOUSE
|
||||||
| θ value | 1.29031e-22 (rank : 37) | NC score | 0.826917 (rank : 36) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P40645, Q62250, Q9QWS5 | Gene names | Sox6, Sox-6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6 (SOX-LZ). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 1.6852e-22 (rank : 38) | NC score | 0.822201 (rank : 37) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SOX30_MOUSE
|
||||||
| θ value | 2.69047e-20 (rank : 39) | NC score | 0.818779 (rank : 39) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SOX30_HUMAN
|
||||||
| θ value | 1.47631e-18 (rank : 40) | NC score | 0.812464 (rank : 40) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
CIC_MOUSE
|
||||||
| θ value | 8.67504e-11 (rank : 41) | NC score | 0.659257 (rank : 43) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CIC_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 42) | NC score | 0.620343 (rank : 47) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
BBX_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 43) | NC score | 0.666455 (rank : 41) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8WY36, Q2TAJ1, Q7L3J8, Q7LBY8, Q8NDB0, Q8WY35, Q9H0J6 | Gene names | BBX, HBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
BBX_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 44) | NC score | 0.661922 (rank : 42) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8VBW5, Q3TZK1, Q6NXY8, Q6PEU3, Q8BQJ7, Q8C7E0, Q8CDQ0, Q8CDV1, Q8VI48, Q8VI49, Q8VI50, Q9CS94 | Gene names | Bbx, Hbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
TF7L2_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 45) | NC score | 0.593148 (rank : 54) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9NQB0, O00185, Q9NQB1, Q9NQB2, Q9NQB3, Q9NQB4, Q9NQB5, Q9NQB6, Q9NQB7, Q9ULC2 | Gene names | TCF7L2, TCF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (hTCF-4). | |||||
|
TF7L2_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 46) | NC score | 0.595939 (rank : 52) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q924A0, O70574, Q91XP2, Q91XP3, Q91XP4, Q924A1, Q9Z0V3, Q9Z0V4 | Gene names | Tcf7l2, Tcf4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (mTCF-4). | |||||
|
LEF1_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 47) | NC score | 0.593599 (rank : 53) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UJU2, Q9HAZ0 | Gene names | LEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1) (T cell-specific transcription factor 1-alpha) (TCF1-alpha). | |||||
|
LEF1_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 48) | NC score | 0.592588 (rank : 55) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P27782 | Gene names | Lef1, Lef-1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1). | |||||
|
TCF7_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 49) | NC score | 0.646111 (rank : 46) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P36402, Q9UKI4 | Gene names | TCF7, TCF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
TCF7_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 50) | NC score | 0.607139 (rank : 48) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
HM20B_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 51) | NC score | 0.601826 (rank : 50) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9P0W2, Q6IBP8, Q8NBD5, Q9HD21, Q9Y491, Q9Y4A2 | Gene names | HMG20B, BRAF35, HMGX2, SMARCE1R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35) (Sox-like transcriptional factor). | |||||
|
TF7L1_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 52) | NC score | 0.573473 (rank : 57) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Z1J1, O70450, O70573 | Gene names | Tcf7l1, Tcf3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF-3) (mTCF-3). | |||||
|
TF7L1_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 53) | NC score | 0.570009 (rank : 59) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9HCS4, Q53R97, Q6PD70, Q9NP00 | Gene names | TCF7L1, TCF3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF- 3). | |||||
|
UBF1_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 54) | NC score | 0.489382 (rank : 68) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
HMGB3_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 55) | NC score | 0.606476 (rank : 49) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O15347, O95556, Q6NS40 | Gene names | HMGB3, HMG2A, HMG4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
HM20B_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 56) | NC score | 0.580998 (rank : 56) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9Z104, Q9NSF7, Q9NSF8 | Gene names | Hmg20b, Braf35, Hmgx2, Smarce1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35). | |||||
|
HMGB3_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 57) | NC score | 0.598672 (rank : 51) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O54879 | Gene names | Hmgb3, Hmg2a, Hmg4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
HMGB2_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 58) | NC score | 0.548916 (rank : 62) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P30681, Q3UXT1, Q9EQD5 | Gene names | Hmgb2, Hmg2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
HMGB2_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 59) | NC score | 0.558976 (rank : 61) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P26583 | Gene names | HMGB2, HMG2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 60) | NC score | 0.494540 (rank : 67) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
HM20A_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 61) | NC score | 0.499132 (rank : 65) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9NP66, Q53G31, Q9NSF6 | Gene names | HMG20A, HMGX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (HMG domain-containing protein HMGX1). | |||||
|
HM20A_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 62) | NC score | 0.497621 (rank : 66) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9DC33, Q3LSF9, Q6NV87, Q8BSK1, Q8C3C1, Q8CAA0, Q9CYG2 | Gene names | Hmg20a, Ibraf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (iBRAF) (Inhibitor of BRAF35) (HMG domain protein HMGX1). | |||||
|
HBP1_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 63) | NC score | 0.654220 (rank : 45) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O60381, Q8TBM1, Q8TE93, Q96AJ2 | Gene names | HBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
HBP1_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 64) | NC score | 0.654657 (rank : 44) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8R316, Q3V0I4, Q8BUS3, Q8C199 | Gene names | Hbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
SSRP1_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 65) | NC score | 0.563768 (rank : 60) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
SSRP1_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 66) | NC score | 0.571201 (rank : 58) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
SMCE1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 67) | NC score | 0.386549 (rank : 71) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
SMCE1_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 68) | NC score | 0.363035 (rank : 72) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
HMGB1_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 69) | NC score | 0.514829 (rank : 64) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P09429, Q6IBE1 | Gene names | HMGB1, HMG1 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HMGB1_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 70) | NC score | 0.514924 (rank : 63) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P63158, P07155, P27109, P27428 | Gene names | Hmgb1, Hmg-1, Hmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
PB1_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 71) | NC score | 0.300686 (rank : 75) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
TOX_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 72) | NC score | 0.267902 (rank : 79) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O94900, Q96AV5 | Gene names | TOX, KIAA0808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
TOX_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 73) | NC score | 0.269037 (rank : 78) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q66JW3, Q8BKH9, Q8BYQ5, Q8R4H0 | Gene names | Tox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
HMG1X_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 74) | NC score | 0.470386 (rank : 69) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UGV6 | Gene names | HMG1L10 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 1-like 10 (HMG-1L10). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 75) | NC score | 0.036549 (rank : 87) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
CN092_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 76) | NC score | 0.295207 (rank : 76) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O94842 | Gene names | C14orf92, KIAA0737 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
CN092_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 77) | NC score | 0.288701 (rank : 77) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
PHLA1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 78) | NC score | 0.043995 (rank : 84) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62392, Q3TY48, Q3UG87 | Gene names | Phlda1, Tdag51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Proline- and glutamine-rich protein) (PQR protein). | |||||
|
HM2L1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 79) | NC score | 0.450483 (rank : 70) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UGU5, O75672, O75673, Q9UMT5 | Gene names | HMG2L1, HMGBCG | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 2-like 1 (Protein HMGBCG). | |||||
|
BTBDC_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 80) | NC score | 0.046055 (rank : 83) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
HXD3_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 81) | NC score | 0.011513 (rank : 117) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P09027, Q3UUD3 | Gene names | Hoxd3, Hox-4.1, Hoxd-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4.1) (Homeobox protein MH-19). | |||||
|
TFAM_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 82) | NC score | 0.327369 (rank : 74) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q00059 | Gene names | TFAM, TCF6L2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Mitochondrial transcription factor 1) (MtTF1) (Transcription factor 6-like 2). | |||||
|
COIA1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 83) | NC score | 0.014906 (rank : 109) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 0.125558 (rank : 84) | NC score | 0.026172 (rank : 94) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
PMS1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 85) | NC score | 0.265145 (rank : 81) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
GTSE1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 86) | NC score | 0.030690 (rank : 89) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYZ3, Q53GX5, Q5R3I6, Q6DHX4, Q9BRE0, Q9UGZ9, Q9Y557 | Gene names | GTSE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (B99 homolog). | |||||
|
HXD3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 87) | NC score | 0.011532 (rank : 116) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P31249, Q99955, Q9BSC5 | Gene names | HOXD3, HOX4A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4A). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.365318 (rank : 88) | NC score | 0.020207 (rank : 99) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
GCX1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 89) | NC score | 0.223409 (rank : 82) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96NM4, Q5TE33, Q5TE34, Q5TE35, Q96IC9, Q9BQN5 | Gene names | GCX1, C20orf100 | |||
|
Domain Architecture |

|
|||||
| Description | Granulosa cell HMG box protein 1 (GCX-1). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 90) | NC score | 0.037789 (rank : 86) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 91) | NC score | 0.025178 (rank : 95) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
GATA6_HUMAN
|
||||||
| θ value | 0.62314 (rank : 92) | NC score | 0.021704 (rank : 97) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92908, P78327 | Gene names | GATA6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
KS6B2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 93) | NC score | -0.005514 (rank : 141) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1M4 | Gene names | Rps6kb2 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosomal protein S6 kinase beta-2 (EC 2.7.11.1) (S6K-beta 2) (70 kDa ribosomal protein S6 kinase 2) (p70-S6KB) (p70 ribosomal S6 kinase beta) (p70 S6Kbeta) (S6K2). | |||||
|
ZDHC5_MOUSE
|
||||||
| θ value | 0.62314 (rank : 94) | NC score | 0.013267 (rank : 112) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VDZ4, Q69ZB5, Q8R2X7 | Gene names | Zdhhc5, Kiaa1748 | |||
|
Domain Architecture |
|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 1.06291 (rank : 95) | NC score | 0.019923 (rank : 101) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
SETBP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 96) | NC score | 0.022703 (rank : 96) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 97) | NC score | 0.028422 (rank : 90) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
CEBPA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 98) | NC score | 0.013924 (rank : 110) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49715, P78319 | Gene names | CEBPA | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
CS007_MOUSE
|
||||||
| θ value | 1.81305 (rank : 99) | NC score | 0.017602 (rank : 103) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
FARP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 100) | NC score | 0.003581 (rank : 135) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 101) | NC score | 0.018170 (rank : 102) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
FIGN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 102) | NC score | 0.005992 (rank : 126) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
HXB3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 103) | NC score | 0.020008 (rank : 100) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P14651, O95615, P17484 | Gene names | HOXB3, HOX2G | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2G) (Hox-2.7). | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 104) | NC score | 0.021280 (rank : 98) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
TBX4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 105) | NC score | 0.007952 (rank : 124) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P57082 | Gene names | TBX4 | |||
|
Domain Architecture |
|
|||||
| Description | T-box transcription factor TBX4 (T-box protein 4). | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 2.36792 (rank : 106) | NC score | 0.004087 (rank : 134) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
ALEX_MOUSE
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.015775 (rank : 108) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
FOXJ2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.008149 (rank : 123) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9P0K8, Q96PS9, Q9NSN5 | Gene names | FOXJ2, FHX | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
ICAM3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.013008 (rank : 113) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32942 | Gene names | ICAM3 | |||
|
Domain Architecture |

|
|||||
| Description | Intercellular adhesion molecule 3 precursor (ICAM-3) (ICAM-R) (CDw50) (CD50 antigen). | |||||
|
TFAM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | 0.361202 (rank : 73) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P40630, P97894, P97906, Q543I8, Q9DBM9 | Gene names | Tfam, Hmgts | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Testis- specific high mobility group protein) (TS-HMG). | |||||
|
ZDHC5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.011098 (rank : 118) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9C0B5, Q6ZMF0, Q8TAK8, Q9H923, Q9UFI7 | Gene names | ZDHHC5, KIAA1748, ZNF375 | |||
|
Domain Architecture |
|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5) (Zinc finger protein 375). | |||||
|
BCAR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.016614 (rank : 105) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.005143 (rank : 130) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CIKS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.016499 (rank : 106) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43734, Q5R3A3, Q9H5W2, Q9H6Y3, Q9NS14, Q9UG72 | Gene names | TRAF3IP2, C6orf4, C6orf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adapter protein CIKS (Connection to IKK and SAPK/JNK) (TRAF3- interacting protein 2) (Nuclear factor NF-kappa-B activator 1) (ACT1). | |||||
|
PTN12_MOUSE
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.005178 (rank : 129) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
BCL3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | -0.000843 (rank : 138) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
FZD7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.002560 (rank : 137) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61090 | Gene names | Fzd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Frizzled-7 precursor (Fz-7) (mFz7). | |||||
|
K1H5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | -0.001319 (rank : 139) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92764, O76012, Q92651 | Gene names | KRTHA5, HHA5, HKA5 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cuticular Ha5 (Hair keratin, type I Ha5). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.008589 (rank : 121) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
PF21B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.008408 (rank : 122) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
SAPS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.006023 (rank : 125) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.026195 (rank : 93) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.035522 (rank : 88) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
DEAF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.012282 (rank : 115) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
DEAF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.012385 (rank : 114) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1T5 | Gene names | Deaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR). | |||||
|
IRS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.013338 (rank : 111) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
IRX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.005020 (rank : 131) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78414, Q7Z2F8, Q8N312 | Gene names | IRX1, IRXA1 | |||
|
Domain Architecture |
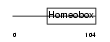
|
|||||
| Description | Iroquois-class homeodomain protein IRX-1 (Iroquois homeobox protein 1) (Homeodomain protein IRXA1). | |||||
|
PI5PA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.009761 (rank : 120) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.004404 (rank : 133) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.015882 (rank : 107) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
SMAD9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.002727 (rank : 136) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15198, O14989 | Gene names | SMAD9, MADH6, MADH9 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Madh6). | |||||
|
TNR7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.005427 (rank : 127) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41272 | Gene names | Tnfrsf7, Cd27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 7 precursor (CD27L receptor) (T-cell activation antigen CD27). | |||||
|
ATX2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.010577 (rank : 119) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.017538 (rank : 104) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
FA5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.004909 (rank : 132) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
FOXJ3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.005212 (rank : 128) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPW0, Q9NSS7 | Gene names | FOXJ3, KIAA1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
GGN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.027373 (rank : 92) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
RERE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.040259 (rank : 85) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
ROBO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | -0.003148 (rank : 140) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6N7, Q7Z300, Q9BUS7 | Gene names | ROBO1, DUTT1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor (H-Robo-1) (Deleted in U twenty twenty). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.027477 (rank : 91) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SP100_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.266493 (rank : 80) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P23497, O75450, Q13343, Q96F70, Q96T24, Q96T95, Q9NP33, Q9UE32 | Gene names | SP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein) (Lysp100b). | |||||
|
SOX7_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
SOX7_MOUSE
|
||||||
| NC score | 0.993655 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P40646, Q9R1T6 | Gene names | Sox7, Sox-7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7 (mSOX7). | |||||
|
SOX18_MOUSE
|
||||||
| NC score | 0.970597 (rank : 3) | θ value | 8.00737e-57 (rank : 5) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P43680, Q9EQ73 | Gene names | Sox18, Sox-18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
SOX18_HUMAN
|
||||||
| NC score | 0.968104 (rank : 4) | θ value | 1.78386e-56 (rank : 6) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P35713, Q9NPH8 | Gene names | SOX18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
SOX17_HUMAN
|
||||||
| NC score | 0.967026 (rank : 5) | θ value | 3.47335e-60 (rank : 3) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9H6I2 | Gene names | SOX17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX17_MOUSE
|
||||||
| NC score | 0.954851 (rank : 6) | θ value | 1.23536e-57 (rank : 4) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q61473, Q61472, Q62248 | Gene names | Sox17, Sox-17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX12_MOUSE
|
||||||
| NC score | 0.945892 (rank : 7) | θ value | 1.62847e-25 (rank : 24) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q04890, P70417, Q6NXL2 | Gene names | Sox12, Sox-12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SOX-12 protein. | |||||
|
SOX12_HUMAN
|
||||||
| NC score | 0.943009 (rank : 8) | θ value | 2.77775e-25 (rank : 25) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O15370, Q9NUD4 | Gene names | SOX12, SOX22 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-12 protein (SOX-22 protein). | |||||
|
SRY_HUMAN
|
||||||
| NC score | 0.940027 (rank : 9) | θ value | 5.79196e-23 (rank : 34) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q05066 | Gene names | SRY, TDF | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
SOX11_HUMAN
|
||||||
| NC score | 0.936859 (rank : 10) | θ value | 8.63488e-27 (rank : 11) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P35716 | Gene names | SOX11 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX15_MOUSE
|
||||||
| NC score | 0.934405 (rank : 11) | θ value | 3.07116e-24 (rank : 30) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P43267, O70204, P70418, Q62246, Q91V00, Q91V43, Q920T1, Q9JLG2 | Gene names | Sox15, Sox-15 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein. | |||||
|
SOX11_MOUSE
|
||||||
| NC score | 0.933039 (rank : 12) | θ value | 2.51237e-26 (rank : 14) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q7M6Y2, O35178, O89036, Q04889, Q80XF0 | Gene names | Sox11, Sox-11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX21_MOUSE
|
||||||
| NC score | 0.932808 (rank : 13) | θ value | 1.05554e-24 (rank : 29) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q811W0, Q8VH35 | Gene names | Sox21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-21. | |||||
|
SOX21_HUMAN
|
||||||
| NC score | 0.932730 (rank : 14) | θ value | 1.05554e-24 (rank : 28) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9Y651, P35715, Q15504 | Gene names | SOX21, SOX25, SOXA | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-21 (SOX-A). | |||||
|
SOX4_HUMAN
|
||||||
| NC score | 0.931224 (rank : 15) | θ value | 4.73814e-25 (rank : 27) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q06945 | Gene names | SOX4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX15_HUMAN
|
||||||
| NC score | 0.930373 (rank : 16) | θ value | 7.30988e-26 (rank : 22) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O60248, P35717, Q9Y6W7 | Gene names | SOX15, SOX12, SOX20, SOX26, SOX27 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein (SOX-20 protein) (SOX-12 protein). | |||||
|
SOX4_MOUSE
|
||||||
| NC score | 0.928558 (rank : 17) | θ value | 3.62785e-25 (rank : 26) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q06831 | Gene names | Sox4, Sox-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX14_MOUSE
|
||||||
| NC score | 0.927618 (rank : 18) | θ value | 5.59698e-26 (rank : 19) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q04892, Q9JLC9, Q9JLD0, Q9QXU4 | Gene names | Sox14, Sox-14 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX14_HUMAN
|
||||||
| NC score | 0.927462 (rank : 19) | θ value | 7.30988e-26 (rank : 21) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | O95416, Q3KPH7 | Gene names | SOX14, SOX28 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX1_HUMAN
|
||||||
| NC score | 0.927296 (rank : 20) | θ value | 3.87602e-27 (rank : 8) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | O00570 | Gene names | SOX1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX1_MOUSE
|
||||||
| NC score | 0.927013 (rank : 21) | θ value | 1.57365e-28 (rank : 7) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P53783 | Gene names | Sox1, Sox-1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX3_MOUSE
|
||||||
| NC score | 0.925439 (rank : 22) | θ value | 2.51237e-26 (rank : 16) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P53784 | Gene names | Sox3, Sox-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SOX2_MOUSE
|
||||||
| NC score | 0.924085 (rank : 23) | θ value | 2.51237e-26 (rank : 15) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P48432 | Gene names | Sox2, Sox-2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX2_HUMAN
|
||||||
| NC score | 0.923775 (rank : 24) | θ value | 5.59698e-26 (rank : 20) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P48431, Q14537 | Gene names | SOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX3_HUMAN
|
||||||
| NC score | 0.921313 (rank : 25) | θ value | 4.28545e-26 (rank : 18) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P41225, P35714, Q9NP49 | Gene names | SOX3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SRY_MOUSE
|
||||||
| NC score | 0.919150 (rank : 26) | θ value | 2.59989e-23 (rank : 33) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q05738 | Gene names | Sry, Tdf, Tdy | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
SOX8_HUMAN
|
||||||
| NC score | 0.915241 (rank : 27) | θ value | 3.28125e-26 (rank : 17) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P57073, Q9NZW2 | Gene names | SOX8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX8_MOUSE
|
||||||
| NC score | 0.913719 (rank : 28) | θ value | 7.30988e-26 (rank : 23) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q04886 | Gene names | Sox8, Sox-8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX10_MOUSE
|
||||||
| NC score | 0.896002 (rank : 29) | θ value | 2.51237e-26 (rank : 13) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q04888, O08518, O09141, O54856, P70416 | Gene names | Sox10, Sox-10, Sox21 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10 (SOX-21) (Transcription factor SOX-M). | |||||
|
SOX10_HUMAN
|
||||||
| NC score | 0.895674 (rank : 30) | θ value | 2.51237e-26 (rank : 12) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
SOX9_MOUSE
|
||||||
| NC score | 0.893738 (rank : 31) | θ value | 6.61148e-27 (rank : 10) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q04887, Q8C7L2, Q91ZK2, Q99KQ0 | Gene names | Sox9, Sox-9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX9_HUMAN
|
||||||
| NC score | 0.892600 (rank : 32) | θ value | 6.61148e-27 (rank : 9) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX5_MOUSE
|
||||||
| NC score | 0.849123 (rank : 33) | θ value | 6.84181e-24 (rank : 32) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P35710, O88184, O89018 | Gene names | Sox5, Sox-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX5_HUMAN
|
||||||
| NC score | 0.846493 (rank : 34) | θ value | 6.84181e-24 (rank : 31) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX6_HUMAN
|
||||||
| NC score | 0.827922 (rank : 35) | θ value | 1.29031e-22 (rank : 36) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P35712, Q9BXQ3, Q9BXQ4, Q9BXQ5, Q9H0I8 | Gene names | SOX6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6. | |||||
|
SOX6_MOUSE
|
||||||
| NC score | 0.826917 (rank : 36) | θ value | 1.29031e-22 (rank : 37) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P40645, Q62250, Q9QWS5 | Gene names | Sox6, Sox-6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6 (SOX-LZ). | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.822201 (rank : 37) | θ value | 1.6852e-22 (rank : 38) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SOX13_HUMAN
|
||||||
| NC score | 0.821028 (rank : 38) | θ value | 1.29031e-22 (rank : 35) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
SOX30_MOUSE
|
||||||
| NC score | 0.818779 (rank : 39) | θ value | 2.69047e-20 (rank : 39) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SOX30_HUMAN
|
||||||
| NC score | 0.812464 (rank : 40) | θ value | 1.47631e-18 (rank : 40) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
BBX_HUMAN
|
||||||
| NC score | 0.666455 (rank : 41) | θ value | 2.13673e-09 (rank : 43) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8WY36, Q2TAJ1, Q7L3J8, Q7LBY8, Q8NDB0, Q8WY35, Q9H0J6 | Gene names | BBX, HBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
BBX_MOUSE
|
||||||
| NC score | 0.661922 (rank : 42) | θ value | 4.76016e-09 (rank : 44) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8VBW5, Q3TZK1, Q6NXY8, Q6PEU3, Q8BQJ7, Q8C7E0, Q8CDQ0, Q8CDV1, Q8VI48, Q8VI49, Q8VI50, Q9CS94 | Gene names | Bbx, Hbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.659257 (rank : 43) | θ value | 8.67504e-11 (rank : 41) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
HBP1_MOUSE
|
||||||
| NC score | 0.654657 (rank : 44) | θ value | 7.1131e-05 (rank : 64) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8R316, Q3V0I4, Q8BUS3, Q8C199 | Gene names | Hbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
HBP1_HUMAN
|
||||||
| NC score | 0.654220 (rank : 45) | θ value | 7.1131e-05 (rank : 63) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O60381, Q8TBM1, Q8TE93, Q96AJ2 | Gene names | HBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
TCF7_HUMAN
|
||||||
| NC score | 0.646111 (rank : 46) | θ value | 2.61198e-07 (rank : 49) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P36402, Q9UKI4 | Gene names | TCF7, TCF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.620343 (rank : 47) | θ value | 4.30538e-10 (rank : 42) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
TCF7_MOUSE
|
||||||
| NC score | 0.607139 (rank : 48) | θ value | 2.61198e-07 (rank : 50) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
HMGB3_HUMAN
|
||||||
| NC score | 0.606476 (rank : 49) | θ value | 4.92598e-06 (rank : 55) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O15347, O95556, Q6NS40 | Gene names | HMGB3, HMG2A, HMG4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
HM20B_HUMAN
|
||||||
| NC score | 0.601826 (rank : 50) | θ value | 5.81887e-07 (rank : 51) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9P0W2, Q6IBP8, Q8NBD5, Q9HD21, Q9Y491, Q9Y4A2 | Gene names | HMG20B, BRAF35, HMGX2, SMARCE1R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35) (Sox-like transcriptional factor). | |||||
|
HMGB3_MOUSE
|
||||||
| NC score | 0.598672 (rank : 51) | θ value | 6.43352e-06 (rank : 57) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O54879 | Gene names | Hmgb3, Hmg2a, Hmg4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
TF7L2_MOUSE
|
||||||
| NC score | 0.595939 (rank : 52) | θ value | 6.87365e-08 (rank : 46) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q924A0, O70574, Q91XP2, Q91XP3, Q91XP4, Q924A1, Q9Z0V3, Q9Z0V4 | Gene names | Tcf7l2, Tcf4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (mTCF-4). | |||||
|
LEF1_HUMAN
|
||||||
| NC score | 0.593599 (rank : 53) | θ value | 1.53129e-07 (rank : 47) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UJU2, Q9HAZ0 | Gene names | LEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1) (T cell-specific transcription factor 1-alpha) (TCF1-alpha). | |||||
|
TF7L2_HUMAN
|
||||||
| NC score | 0.593148 (rank : 54) | θ value | 6.87365e-08 (rank : 45) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9NQB0, O00185, Q9NQB1, Q9NQB2, Q9NQB3, Q9NQB4, Q9NQB5, Q9NQB6, Q9NQB7, Q9ULC2 | Gene names | TCF7L2, TCF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (hTCF-4). | |||||
|
LEF1_MOUSE
|
||||||
| NC score | 0.592588 (rank : 55) | θ value | 1.53129e-07 (rank : 48) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P27782 | Gene names | Lef1, Lef-1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1). | |||||
|
HM20B_MOUSE
|
||||||
| NC score | 0.580998 (rank : 56) | θ value | 6.43352e-06 (rank : 56) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9Z104, Q9NSF7, Q9NSF8 | Gene names | Hmg20b, Braf35, Hmgx2, Smarce1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35). | |||||
|
TF7L1_MOUSE
|
||||||
| NC score | 0.573473 (rank : 57) | θ value | 5.81887e-07 (rank : 52) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Z1J1, O70450, O70573 | Gene names | Tcf7l1, Tcf3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF-3) (mTCF-3). | |||||
|
SSRP1_MOUSE
|
||||||
| NC score | 0.571201 (rank : 58) | θ value | 9.29e-05 (rank : 66) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
TF7L1_HUMAN
|
||||||
| NC score | 0.570009 (rank : 59) | θ value | 1.29631e-06 (rank : 53) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9HCS4, Q53R97, Q6PD70, Q9NP00 | Gene names | TCF7L1, TCF3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF- 3). | |||||
|
SSRP1_HUMAN
|
||||||
| NC score | 0.563768 (rank : 60) | θ value | 9.29e-05 (rank : 65) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
HMGB2_HUMAN
|
||||||
| NC score | 0.558976 (rank : 61) | θ value | 1.09739e-05 (rank : 59) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P26583 | Gene names | HMGB2, HMG2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
HMGB2_MOUSE
|
||||||
| NC score | 0.548916 (rank : 62) | θ value | 8.40245e-06 (rank : 58) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P30681, Q3UXT1, Q9EQD5 | Gene names | Hmgb2, Hmg2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
HMGB1_MOUSE
|
||||||
| NC score | 0.514924 (rank : 63) | θ value | 0.000270298 (rank : 70) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P63158, P07155, P27109, P27428 | Gene names | Hmgb1, Hmg-1, Hmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HMGB1_HUMAN
|
||||||
| NC score | 0.514829 (rank : 64) | θ value | 0.000270298 (rank : 69) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P09429, Q6IBE1 | Gene names | HMGB1, HMG1 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HM20A_HUMAN
|
||||||
| NC score | 0.499132 (rank : 65) | θ value | 4.1701e-05 (rank : 61) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9NP66, Q53G31, Q9NSF6 | Gene names | HMG20A, HMGX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (HMG domain-containing protein HMGX1). | |||||
|
HM20A_MOUSE
|
||||||
| NC score | 0.497621 (rank : 66) | θ value | 4.1701e-05 (rank : 62) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9DC33, Q3LSF9, Q6NV87, Q8BSK1, Q8C3C1, Q8CAA0, Q9CYG2 | Gene names | Hmg20a, Ibraf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (iBRAF) (Inhibitor of BRAF35) (HMG domain protein HMGX1). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.494540 (rank : 67) | θ value | 1.09739e-05 (rank : 60) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
UBF1_MOUSE
|
||||||
| NC score | 0.489382 (rank : 68) | θ value | 3.77169e-06 (rank : 54) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
HMG1X_HUMAN
|
||||||
| NC score | 0.470386 (rank : 69) | θ value | 0.00665767 (rank : 74) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UGV6 | Gene names | HMG1L10 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 1-like 10 (HMG-1L10). | |||||
|
HM2L1_HUMAN
|
||||||
| NC score | 0.450483 (rank : 70) | θ value | 0.0252991 (rank : 79) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UGU5, O75672, O75673, Q9UMT5 | Gene names | HMG2L1, HMGBCG | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 2-like 1 (Protein HMGBCG). | |||||
|
SMCE1_HUMAN
|
||||||
| NC score | 0.386549 (rank : 71) | θ value | 0.000121331 (rank : 67) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
SMCE1_MOUSE
|
||||||
| NC score | 0.363035 (rank : 72) | θ value | 0.000121331 (rank : 68) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
TFAM_MOUSE
|
||||||
| NC score | 0.361202 (rank : 73) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P40630, P97894, P97906, Q543I8, Q9DBM9 | Gene names | Tfam, Hmgts | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Testis- specific high mobility group protein) (TS-HMG). | |||||
|
TFAM_HUMAN
|
||||||
| NC score | 0.327369 (rank : 74) | θ value | 0.0736092 (rank : 82) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q00059 | Gene names | TFAM, TCF6L2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Mitochondrial transcription factor 1) (MtTF1) (Transcription factor 6-like 2). | |||||
|
PB1_HUMAN
|
||||||
| NC score | 0.300686 (rank : 75) | θ value | 0.00175202 (rank : 71) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
CN092_HUMAN
|
||||||
| NC score | 0.295207 (rank : 76) | θ value | 0.0113563 (rank : 76) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O94842 | Gene names | C14orf92, KIAA0737 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
CN092_MOUSE
|
||||||
| NC score | 0.288701 (rank : 77) | θ value | 0.0113563 (rank : 77) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
TOX_MOUSE
|
||||||
| NC score | 0.269037 (rank : 78) | θ value | 0.00509761 (rank : 73) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q66JW3, Q8BKH9, Q8BYQ5, Q8R4H0 | Gene names | Tox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
TOX_HUMAN
|
||||||
| NC score | 0.267902 (rank : 79) | θ value | 0.00509761 (rank : 72) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O94900, Q96AV5 | Gene names | TOX, KIAA0808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
SP100_HUMAN
|
||||||
| NC score | 0.266493 (rank : 80) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P23497, O75450, Q13343, Q96F70, Q96T24, Q96T95, Q9NP33, Q9UE32 | Gene names | SP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein) (Lysp100b). | |||||
|
PMS1_HUMAN
|
||||||
| NC score | 0.265145 (rank : 81) | θ value | 0.163984 (rank : 85) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
GCX1_HUMAN
|
||||||
| NC score | 0.223409 (rank : 82) | θ value | 0.47712 (rank : 89) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96NM4, Q5TE33, Q5TE34, Q5TE35, Q96IC9, Q9BQN5 | Gene names | GCX1, C20orf100 | |||
|
Domain Architecture |

|
|||||
| Description | Granulosa cell HMG box protein 1 (GCX-1). | |||||
|
BTBDC_HUMAN
|
||||||
| NC score | 0.046055 (rank : 83) | θ value | 0.0736092 (rank : 80) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
PHLA1_MOUSE
|
||||||
| NC score | 0.043995 (rank : 84) | θ value | 0.0113563 (rank : 78) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62392, Q3TY48, Q3UG87 | Gene names | Phlda1, Tdag51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Proline- and glutamine-rich protein) (PQR protein). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.040259 (rank : 85) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.037789 (rank : 86) | θ value | 0.47712 (rank : 90) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.036549 (rank : 87) | θ value | 0.00665767 (rank : 75) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.035522 (rank : 88) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
GTSE1_HUMAN
|
||||||
| NC score | 0.030690 (rank : 89) | θ value | 0.21417 (rank : 86) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYZ3, Q53GX5, Q5R3I6, Q6DHX4, Q9BRE0, Q9UGZ9, Q9Y557 | Gene names | GTSE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (B99 homolog). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.028422 (rank : 90) | θ value | 1.38821 (rank : 97) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.027477 (rank : 91) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.027373 (rank : 92) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.026195 (rank : 93) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.026172 (rank : 94) | θ value | 0.125558 (rank : 84) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.025178 (rank : 95) | θ value | 0.47712 (rank : 91) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SETBP_MOUSE
|
||||||
| NC score | 0.022703 (rank : 96) | θ value | 1.06291 (rank : 96) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
GATA6_HUMAN
|
||||||
| NC score | 0.021704 (rank : 97) | θ value | 0.62314 (rank : 92) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92908, P78327 | Gene names | GATA6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.021280 (rank : 98) | θ value | 2.36792 (rank : 104) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.020207 (rank : 99) | θ value | 0.365318 (rank : 88) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
HXB3_HUMAN
|
||||||
| NC score | 0.020008 (rank : 100) | θ value | 2.36792 (rank : 103) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P14651, O95615, P17484 | Gene names | HOXB3, HOX2G | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2G) (Hox-2.7). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.019923 (rank : 101) | θ value | 1.06291 (rank : 95) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.018170 (rank : 102) | θ value | 1.81305 (rank : 101) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.017602 (rank : 103) | θ value | 1.81305 (rank : 99) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.017538 (rank : 104) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
BCAR1_HUMAN
|
||||||
| NC score | 0.016614 (rank : 105) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
CIKS_HUMAN
|
||||||
| NC score | 0.016499 (rank : 106) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43734, Q5R3A3, Q9H5W2, Q9H6Y3, Q9NS14, Q9UG72 | Gene names | TRAF3IP2, C6orf4, C6orf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adapter protein CIKS (Connection to IKK and SAPK/JNK) (TRAF3- interacting protein 2) (Nuclear factor NF-kappa-B activator 1) (ACT1). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.015882 (rank : 107) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
ALEX_MOUSE
|
||||||
| NC score | 0.015775 (rank : 108) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
COIA1_MOUSE
|
||||||
| NC score | 0.014906 (rank : 109) | θ value | 0.0961366 (rank : 83) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
CEBPA_HUMAN
|
||||||
| NC score | 0.013924 (rank : 110) | θ value | 1.81305 (rank : 98) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49715, P78319 | Gene names | CEBPA | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
IRS2_HUMAN
|
||||||
| NC score | 0.013338 (rank : 111) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
ZDHC5_MOUSE
|
||||||
| NC score | 0.013267 (rank : 112) | θ value | 0.62314 (rank : 94) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VDZ4, Q69ZB5, Q8R2X7 | Gene names | Zdhhc5, Kiaa1748 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5). | |||||
|
ICAM3_HUMAN
|
||||||
| NC score | 0.013008 (rank : 113) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32942 | Gene names | ICAM3 | |||
|
Domain Architecture |

|
|||||
| Description | Intercellular adhesion molecule 3 precursor (ICAM-3) (ICAM-R) (CDw50) (CD50 antigen). | |||||
|
DEAF1_MOUSE
|
||||||
| NC score | 0.012385 (rank : 114) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1T5 | Gene names | Deaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR). | |||||
|
DEAF1_HUMAN
|
||||||
| NC score | 0.012282 (rank : 115) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
HXD3_HUMAN
|
||||||
| NC score | 0.011532 (rank : 116) | θ value | 0.21417 (rank : 87) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P31249, Q99955, Q9BSC5 | Gene names | HOXD3, HOX4A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4A). | |||||
|
HXD3_MOUSE
|
||||||
| NC score | 0.011513 (rank : 117) | θ value | 0.0736092 (rank : 81) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P09027, Q3UUD3 | Gene names | Hoxd3, Hox-4.1, Hoxd-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4.1) (Homeobox protein MH-19). | |||||
|
ZDHC5_HUMAN
|
||||||
| NC score | 0.011098 (rank : 118) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9C0B5, Q6ZMF0, Q8TAK8, Q9H923, Q9UFI7 | Gene names | ZDHHC5, KIAA1748, ZNF375 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5) (Zinc finger protein 375). | |||||
|
ATX2_HUMAN
|
||||||
| NC score | 0.010577 (rank : 119) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
PI5PA_MOUSE
|
||||||
| NC score | 0.009761 (rank : 120) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.008589 (rank : 121) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
PF21B_HUMAN
|
||||||
| NC score | 0.008408 (rank : 122) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
FOXJ2_HUMAN
|
||||||
| NC score | 0.008149 (rank : 123) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9P0K8, Q96PS9, Q9NSN5 | Gene names | FOXJ2, FHX | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
TBX4_HUMAN
|
||||||
| NC score | 0.007952 (rank : 124) | θ value | 2.36792 (rank : 105) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P57082 | Gene names | TBX4 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX4 (T-box protein 4). | |||||
|
SAPS1_MOUSE
|
||||||
| NC score | 0.006023 (rank : 125) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
FIGN_HUMAN
|
||||||
| NC score | 0.005992 (rank : 126) | θ value | 2.36792 (rank : 102) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
TNR7_MOUSE
|
||||||
| NC score | 0.005427 (rank : 127) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41272 | Gene names | Tnfrsf7, Cd27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 7 precursor (CD27L receptor) (T-cell activation antigen CD27). | |||||
|
FOXJ3_HUMAN
|
||||||
| NC score | 0.005212 (rank : 128) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPW0, Q9NSS7 | Gene names | FOXJ3, KIAA1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
PTN12_MOUSE
|
||||||
| NC score | 0.005178 (rank : 129) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.005143 (rank : 130) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
IRX1_HUMAN
|
||||||
| NC score | 0.005020 (rank : 131) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78414, Q7Z2F8, Q8N312 | Gene names | IRX1, IRXA1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-1 (Iroquois homeobox protein 1) (Homeodomain protein IRXA1). | |||||
|
FA5_HUMAN
|
||||||
| NC score | 0.004909 (rank : 132) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.004404 (rank : 133) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.004087 (rank : 134) | θ value | 2.36792 (rank : 106) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
FARP2_HUMAN
|
||||||
| NC score | 0.003581 (rank : 135) | θ value | 1.81305 (rank : 100) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
SMAD9_HUMAN
|
||||||
| NC score | 0.002727 (rank : 136) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15198, O14989 | Gene names | SMAD9, MADH6, MADH9 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Madh6). | |||||
|
FZD7_MOUSE
|
||||||
| NC score | 0.002560 (rank : 137) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61090 | Gene names | Fzd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Frizzled-7 precursor (Fz-7) (mFz7). | |||||
|
BCL3_MOUSE
|
||||||
| NC score | -0.000843 (rank : 138) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
K1H5_HUMAN
|
||||||
| NC score | -0.001319 (rank : 139) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92764, O76012, Q92651 | Gene names | KRTHA5, HHA5, HKA5 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha5 (Hair keratin, type I Ha5). | |||||
|
ROBO1_HUMAN
|
||||||
| NC score | -0.003148 (rank : 140) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6N7, Q7Z300, Q9BUS7 | Gene names | ROBO1, DUTT1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor (H-Robo-1) (Deleted in U twenty twenty). | |||||
|
KS6B2_MOUSE
|
||||||
| NC score | -0.005514 (rank : 141) | θ value | 0.62314 (rank : 93) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1M4 | Gene names | Rps6kb2 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosomal protein S6 kinase beta-2 (EC 2.7.11.1) (S6K-beta 2) (70 kDa ribosomal protein S6 kinase 2) (p70-S6KB) (p70 ribosomal S6 kinase beta) (p70 S6Kbeta) (S6K2). | |||||