Please be patient as the page loads
|
CEBPA_HUMAN
|
||||||
| SwissProt Accessions | P49715, P78319 | Gene names | CEBPA | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CEBPA_HUMAN
|
||||||
| θ value | 1.06727e-101 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | P49715, P78319 | Gene names | CEBPA | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
CEBPA_MOUSE
|
||||||
| θ value | 1.3501e-96 (rank : 2) | NC score | 0.990707 (rank : 2) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P53566, Q91XB6 | Gene names | Cebpa | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
CEBPE_HUMAN
|
||||||
| θ value | 2.76489e-41 (rank : 3) | NC score | 0.851302 (rank : 6) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
CEBPE_MOUSE
|
||||||
| θ value | 1.28434e-38 (rank : 4) | NC score | 0.890027 (rank : 3) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6PZD9 | Gene names | Cebpe | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
CEBPB_HUMAN
|
||||||
| θ value | 2.68423e-28 (rank : 5) | NC score | 0.874622 (rank : 5) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P17676, Q96IH2, Q9H4Z5 | Gene names | CEBPB, TCF5 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Nuclear factor NF- IL6) (Transcription factor 5). | |||||
|
CEBPB_MOUSE
|
||||||
| θ value | 1.24688e-25 (rank : 6) | NC score | 0.882115 (rank : 4) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P28033 | Gene names | Cebpb | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Interleukin-6- dependent-binding protein) (IL-6DBP) (Liver-enriched transcriptional activator) (LAP) (AGP/EBP). | |||||
|
CEBPD_HUMAN
|
||||||
| θ value | 3.51386e-20 (rank : 7) | NC score | 0.825605 (rank : 7) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P49716, Q14937 | Gene names | CEBPD | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (Nuclear factor NF- IL6-beta) (NF-IL6-beta). | |||||
|
CEBPD_MOUSE
|
||||||
| θ value | 1.47631e-18 (rank : 8) | NC score | 0.824415 (rank : 8) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q00322 | Gene names | Cebpd, Crp3 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (C/EBP-related protein 3). | |||||
|
CEBPG_HUMAN
|
||||||
| θ value | 2.0648e-12 (rank : 9) | NC score | 0.770466 (rank : 10) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P53567, Q5U052 | Gene names | CEBPG | |||
|
Domain Architecture |
|
|||||
| Description | CCAAT/enhancer-binding protein gamma (C/EBP gamma). | |||||
|
CEBPG_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 10) | NC score | 0.775598 (rank : 9) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P53568 | Gene names | Cebpg | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein gamma (C/EBP gamma) (Immunoglobulin enhancer-binding protein 1) (IG/EBP-1) (Granulocyte colony-stimulating factor promoter element 1-binding protein) (GPE1-binding protein) (GPE1-BP). | |||||
|
DDIT3_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 11) | NC score | 0.459199 (rank : 11) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35639, Q91YW9 | Gene names | Ddit3, Chop, Gadd153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
DDIT3_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 12) | NC score | 0.410121 (rank : 12) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35638 | Gene names | DDIT3, CHOP, GADD153 | |||
|
Domain Architecture |

|
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 13) | NC score | 0.044711 (rank : 56) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
HLF_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 14) | NC score | 0.291047 (rank : 19) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q16534 | Gene names | HLF | |||
|
Domain Architecture |

|
|||||
| Description | Hepatic leukemia factor. | |||||
|
HLF_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 15) | NC score | 0.290451 (rank : 20) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BW74, Q6PF83 | Gene names | Hlf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatic leukemia factor. | |||||
|
FOSL2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 16) | NC score | 0.272236 (rank : 22) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P15408 | Gene names | FOSL2, FRA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
FOSL2_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 17) | NC score | 0.273584 (rank : 21) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P47930 | Gene names | Fosl2, Fra-2, Fra2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
ATF3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 18) | NC score | 0.303640 (rank : 15) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q60765 | Gene names | Atf3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3) (Transcription factor LRG-21). | |||||
|
TEF_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 19) | NC score | 0.296013 (rank : 17) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q10587, Q15729, Q7Z3J7, Q8IU94, Q96TG4 | Gene names | TEF | |||
|
Domain Architecture |

|
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
TEF_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 20) | NC score | 0.296172 (rank : 16) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JLC6, Q3U426, Q3UGF4, Q3UM49, Q3URC8, Q6QHT6, Q8C6I0, Q8VD02 | Gene names | Tef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
BATF_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 21) | NC score | 0.364741 (rank : 13) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16520 | Gene names | BATF | |||
|
Domain Architecture |
|
|||||
| Description | ATF-like basic leucine zipper transcriptional factor B-ATF (SF-HT- activated gene 2) (SFA-2). | |||||
|
K2C8_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 22) | NC score | 0.032668 (rank : 66) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P05787, Q14099, Q14716, Q14717, Q53GJ0, Q6DHW5, Q6GMY0, Q96J60 | Gene names | KRT8, CYK8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8). | |||||
|
ATF3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 23) | NC score | 0.294879 (rank : 18) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P18847 | Gene names | ATF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3). | |||||
|
BATF_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 24) | NC score | 0.353513 (rank : 14) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O35284 | Gene names | Batf | |||
|
Domain Architecture |
|
|||||
| Description | ATF-like basic leucine zipper transcription factor B-ATF. | |||||
|
K2C8_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 25) | NC score | 0.031120 (rank : 68) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P11679, Q3KQK5, Q3TGI1, Q3TJE1, Q3TKY7, Q61463, Q61518, Q61519 | Gene names | Krt8, Krt2-8 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8) (Cytokeratin endo A). | |||||
|
FOSL1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 26) | NC score | 0.242905 (rank : 26) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOSL1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 27) | NC score | 0.248668 (rank : 25) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
GCM1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 28) | NC score | 0.066765 (rank : 49) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NP62, Q5T0X0, Q99468, Q9P1X3 | Gene names | GCM1, GCMA | |||
|
Domain Architecture |
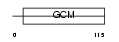
|
|||||
| Description | Chorion-specific transcription factor GCMa (Glial cells missing homolog 1) (GCM motif protein 1) (hGCMa). | |||||
|
ABL1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 29) | NC score | 0.009296 (rank : 104) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
CO4A2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.016682 (rank : 98) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
DBP_MOUSE
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.251878 (rank : 24) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60925, Q8VCX3 | Gene names | Dbp | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein). | |||||
|
KCNH4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.045409 (rank : 55) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.021813 (rank : 88) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
DBP_HUMAN
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.253295 (rank : 23) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q10586 | Gene names | DBP | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein) (TAXREB302). | |||||
|
FOSB_HUMAN
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.235863 (rank : 27) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
FOSB_MOUSE
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.224007 (rank : 28) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
NGN2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.059698 (rank : 53) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70447, P70237 | Gene names | Neurog2, Ath4a, Atoh4, Ngn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 2 (Atonal protein homolog 4) (Helix-loop-helix protein mATH-4A) (MATH4A). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.024194 (rank : 86) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
ATF7_HUMAN
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.135218 (rank : 34) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ATF7_MOUSE
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.135938 (rank : 33) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
CREB5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.156559 (rank : 31) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.031335 (rank : 67) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.029375 (rank : 74) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RU17_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.038276 (rank : 61) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P08621, P78493, P78494, Q15364, Q15686, Q15687, Q15689, Q99377, Q9UE45, Q9UE46, Q9UE47, Q9UE48, Q9UFQ6 | Gene names | SNRP70, RPU1, U1AP1 | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 snRNP 70 kDa) (snRNP70) (U1-70K). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.033532 (rank : 65) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
PER1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.024734 (rank : 83) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.027111 (rank : 78) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
FOS_HUMAN
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.194711 (rank : 30) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P01100, P18849 | Gene names | FOS, G0S7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos) (G0/G1 switch regulatory protein 7). | |||||
|
FOS_MOUSE
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.195287 (rank : 29) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P01101 | Gene names | Fos | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos). | |||||
|
NGN3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.060089 (rank : 51) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70661 | Gene names | Neurog3, Ath4b, Atoh5, Ngn3 | |||
|
Domain Architecture |
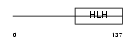
|
|||||
| Description | Neurogenin 3 (Atonal protein homolog 5) (Helix-loop-helix protein mATH-4B) (MATH4B). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.003084 (rank : 114) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.041032 (rank : 57) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
K2027_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.040303 (rank : 58) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.024721 (rank : 84) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.021551 (rank : 89) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
NGN3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.057316 (rank : 54) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4Z2, Q9BY24 | Gene names | NEUROG3, NGN3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3. | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.024556 (rank : 85) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
RU17_MOUSE
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.033738 (rank : 64) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62376, Q3UIW4 | Gene names | Snrp70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 SNRNP 70 kDa) (snRNP70). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.034666 (rank : 63) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
SOX7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.013924 (rank : 101) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
STRAD_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.068388 (rank : 48) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60924 | Gene names | Stra13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-inducible E3 protein (Hematopoietic-specific protein E3). | |||||
|
TCF7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.027172 (rank : 77) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P36402, Q9UKI4 | Gene names | TCF7, TCF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.020795 (rank : 91) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
AMOL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.025080 (rank : 82) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y2J4, Q53EP1, Q96F99, Q9UKB4 | Gene names | AMOTL2, KIAA0989 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2 (Leman coiled-coil protein) (LCCP). | |||||
|
CRSP7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.027226 (rank : 76) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
HXD1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.004633 (rank : 113) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01822 | Gene names | Hoxd1, Hox-4.9, Hoxd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D1 (Hox-4.9). | |||||
|
USH1C_MOUSE
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.026544 (rank : 80) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.025770 (rank : 81) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.010580 (rank : 103) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
LRP5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.006060 (rank : 111) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91VN0, O88883, Q9R208 | Gene names | Lrp5, Lrp7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor (LRP7) (Lr3). | |||||
|
RPA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.014887 (rank : 100) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95602, Q9UEH0, Q9UFT9 | Gene names | POLR1A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
SEN54_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.021002 (rank : 90) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z6J9, Q86WV3, Q86XE4, Q8N9H2 | Gene names | TSEN54, SEN54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen54 (tRNA-intron endonuclease Sen54) (HsSen54). | |||||
|
SFR11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.059837 (rank : 52) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
TR19L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.017078 (rank : 97) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969Z4, Q86V34, Q96JU1, Q9BUX7 | Gene names | TNFRSF19L, RELT | |||
|
Domain Architecture |
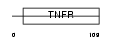
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 19L precursor (Receptor expressed in lymphoid tissues). | |||||
|
ZN318_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.030300 (rank : 70) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
AZI1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.029687 (rank : 73) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.019215 (rank : 93) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
IFT74_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.026937 (rank : 79) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.030192 (rank : 72) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.020039 (rank : 92) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
PF21A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.017639 (rank : 96) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PTPRN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.006316 (rank : 110) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16849, Q08319 | Gene names | PTPRN, ICA3, ICA512 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2) (Islet cell antigen 512) (ICA 512) (Islet cell autoantigen 3). | |||||
|
SGOL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.034951 (rank : 62) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q562F6, Q53RR9, Q53T20, Q86XY4, Q8IWK2, Q8IZK1, Q8N1Q5, Q96LQ3 | Gene names | SGOL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 2 (Tripin). | |||||
|
CHMP6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.030292 (rank : 71) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96FZ7, Q53FU4, Q9HAE8 | Gene names | CHMP6, VPS20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 6 (Chromatin-modifying protein 6) (Vacuolar protein sorting-associated protein 20) (hVps20). | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.010647 (rank : 102) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
MEIS2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.006841 (rank : 109) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97367, O35676, O35677, P97403, P97404 | Gene names | Meis2, Mrg1, Stra10 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Meis2 (Meis1-related protein 1). | |||||
|
PLXB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.005168 (rank : 112) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15031, Q7KZU3, Q9BSU7 | Gene names | PLXNB2, KIAA0315 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B2 precursor (MM1). | |||||
|
SGOL2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.039979 (rank : 59) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TSY8, Q811I4, Q8C9C4, Q8CGJ4, Q9CS55 | Gene names | Sgol2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 2. | |||||
|
SMO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.007143 (rank : 108) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56726 | Gene names | Smo, Smoh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smoothened homolog precursor (SMO). | |||||
|
TJAP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.017896 (rank : 95) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DCD5, Q8CFL7, Q8R5I2 | Gene names | Tjap1, Pilt, Tjp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.008075 (rank : 106) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CHMP6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.031081 (rank : 69) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P0C0A3 | Gene names | Chmp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 6 (Chromatin-modifying protein 6). | |||||
|
DCBD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.007931 (rank : 107) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D4J3, Q8R327, Q9D696 | Gene names | Dcbld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
MAFG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.022532 (rank : 87) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O54790 | Gene names | Mafg | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafG (V-maf musculoaponeurotic fibrosarcoma oncogene homolog G). | |||||
|
MYC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.016488 (rank : 99) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
NGN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.039940 (rank : 60) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H2A3, Q8N416 | Gene names | NEUROG2, NGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogenin 2. | |||||
|
NTBP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.028547 (rank : 75) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BRM2, Q8CAA8 | Gene names | Scyl1bp1, Ntklbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (mNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
TCF7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.017962 (rank : 94) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
TTLL4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.008143 (rank : 105) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80UG8, Q5U4C4 | Gene names | Ttll4, Kiaa0173 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
ATF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.097246 (rank : 39) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
ATF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.097199 (rank : 40) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
ATF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.100799 (rank : 37) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P18848, Q9UH31 | Gene names | ATF4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (Activating transcription factor 4) (DNA-binding protein TAXREB67) (Cyclic AMP response element-binding protein 2) (CREB2). | |||||
|
ATF4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.098921 (rank : 38) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q06507, Q61906 | Gene names | Atf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (C/EBP-related ATF) (C/ATF) (TAXREB67 homolog). | |||||
|
ATF6A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.061615 (rank : 50) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P18850, O15139, Q5VW62, Q6IPB5, Q9UEC9 | Gene names | ATF6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 alpha (Activating transcription factor 6 alpha) (ATF6-alpha). | |||||
|
CREB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.069056 (rank : 47) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43889, O14671, O14919, Q96GK8, Q9H2W3, Q9UE77 | Gene names | CREB3, LZIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Luman protein) (Transcription factor LZIP-alpha). | |||||
|
CREB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.113428 (rank : 36) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61817, Q99M21, Q9CVK9 | Gene names | Creb3, Lzip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Transcription factor LZIP). | |||||
|
JUNB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.089031 (rank : 44) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P17275, Q96GH3 | Gene names | JUNB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-B. | |||||
|
JUNB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.093053 (rank : 41) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P09450, Q8C2G9 | Gene names | Junb, Jun-b | |||
|
Domain Architecture |
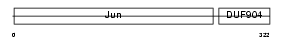
|
|||||
| Description | Transcription factor jun-B. | |||||
|
JUND_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.077275 (rank : 45) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P17535 | Gene names | JUND | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
JUND_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.073378 (rank : 46) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15066 | Gene names | Jund, Jun-d, Jund1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
JUN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.092006 (rank : 42) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05412, Q96G93 | Gene names | JUN | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (p39). | |||||
|
JUN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.091889 (rank : 43) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05627 | Gene names | Jun | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (Jun A) (AH119). | |||||
|
XBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.129398 (rank : 35) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P17861, Q8WYK6, Q969P1, Q96BD7 | Gene names | XBP1, TREB5, XBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5). | |||||
|
XBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.149050 (rank : 32) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35426, Q8VHM0, Q922G5, Q9ESS3 | Gene names | Xbp1, Treb5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5 homolog). | |||||
|
CEBPA_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.06727e-101 (rank : 1) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | P49715, P78319 | Gene names | CEBPA | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
CEBPA_MOUSE
|
||||||
| NC score | 0.990707 (rank : 2) | θ value | 1.3501e-96 (rank : 2) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P53566, Q91XB6 | Gene names | Cebpa | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
CEBPE_MOUSE
|
||||||
| NC score | 0.890027 (rank : 3) | θ value | 1.28434e-38 (rank : 4) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6PZD9 | Gene names | Cebpe | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
CEBPB_MOUSE
|
||||||
| NC score | 0.882115 (rank : 4) | θ value | 1.24688e-25 (rank : 6) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P28033 | Gene names | Cebpb | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Interleukin-6- dependent-binding protein) (IL-6DBP) (Liver-enriched transcriptional activator) (LAP) (AGP/EBP). | |||||
|
CEBPB_HUMAN
|
||||||
| NC score | 0.874622 (rank : 5) | θ value | 2.68423e-28 (rank : 5) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P17676, Q96IH2, Q9H4Z5 | Gene names | CEBPB, TCF5 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Nuclear factor NF- IL6) (Transcription factor 5). | |||||
|
CEBPE_HUMAN
|
||||||
| NC score | 0.851302 (rank : 6) | θ value | 2.76489e-41 (rank : 3) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
CEBPD_HUMAN
|
||||||
| NC score | 0.825605 (rank : 7) | θ value | 3.51386e-20 (rank : 7) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P49716, Q14937 | Gene names | CEBPD | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (Nuclear factor NF- IL6-beta) (NF-IL6-beta). | |||||
|
CEBPD_MOUSE
|
||||||
| NC score | 0.824415 (rank : 8) | θ value | 1.47631e-18 (rank : 8) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q00322 | Gene names | Cebpd, Crp3 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (C/EBP-related protein 3). | |||||
|
CEBPG_MOUSE
|
||||||
| NC score | 0.775598 (rank : 9) | θ value | 4.59992e-12 (rank : 10) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P53568 | Gene names | Cebpg | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein gamma (C/EBP gamma) (Immunoglobulin enhancer-binding protein 1) (IG/EBP-1) (Granulocyte colony-stimulating factor promoter element 1-binding protein) (GPE1-binding protein) (GPE1-BP). | |||||
|
CEBPG_HUMAN
|
||||||
| NC score | 0.770466 (rank : 10) | θ value | 2.0648e-12 (rank : 9) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P53567, Q5U052 | Gene names | CEBPG | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein gamma (C/EBP gamma). | |||||
|
DDIT3_MOUSE
|
||||||
| NC score | 0.459199 (rank : 11) | θ value | 0.000270298 (rank : 11) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35639, Q91YW9 | Gene names | Ddit3, Chop, Gadd153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
DDIT3_HUMAN
|
||||||
| NC score | 0.410121 (rank : 12) | θ value | 0.000786445 (rank : 12) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35638 | Gene names | DDIT3, CHOP, GADD153 | |||
|
Domain Architecture |

|
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
BATF_HUMAN
|
||||||
| NC score | 0.364741 (rank : 13) | θ value | 0.0330416 (rank : 21) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16520 | Gene names | BATF | |||
|
Domain Architecture |

|
|||||
| Description | ATF-like basic leucine zipper transcriptional factor B-ATF (SF-HT- activated gene 2) (SFA-2). | |||||
|
BATF_MOUSE
|
||||||
| NC score | 0.353513 (rank : 14) | θ value | 0.0431538 (rank : 24) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O35284 | Gene names | Batf | |||
|
Domain Architecture |

|
|||||
| Description | ATF-like basic leucine zipper transcription factor B-ATF. | |||||
|
ATF3_MOUSE
|
||||||
| NC score | 0.303640 (rank : 15) | θ value | 0.0252991 (rank : 18) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q60765 | Gene names | Atf3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3) (Transcription factor LRG-21). | |||||
|
TEF_MOUSE
|
||||||
| NC score | 0.296172 (rank : 16) | θ value | 0.0252991 (rank : 20) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JLC6, Q3U426, Q3UGF4, Q3UM49, Q3URC8, Q6QHT6, Q8C6I0, Q8VD02 | Gene names | Tef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
TEF_HUMAN
|
||||||
| NC score | 0.296013 (rank : 17) | θ value | 0.0252991 (rank : 19) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q10587, Q15729, Q7Z3J7, Q8IU94, Q96TG4 | Gene names | TEF | |||
|
Domain Architecture |

|
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
ATF3_HUMAN
|
||||||
| NC score | 0.294879 (rank : 18) | θ value | 0.0431538 (rank : 23) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P18847 | Gene names | ATF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3). | |||||
|
HLF_HUMAN
|
||||||
| NC score | 0.291047 (rank : 19) | θ value | 0.0148317 (rank : 14) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q16534 | Gene names | HLF | |||
|
Domain Architecture |

|
|||||
| Description | Hepatic leukemia factor. | |||||
|
HLF_MOUSE
|
||||||
| NC score | 0.290451 (rank : 20) | θ value | 0.0148317 (rank : 15) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BW74, Q6PF83 | Gene names | Hlf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatic leukemia factor. | |||||
|
FOSL2_MOUSE
|
||||||
| NC score | 0.273584 (rank : 21) | θ value | 0.0193708 (rank : 17) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P47930 | Gene names | Fosl2, Fra-2, Fra2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
FOSL2_HUMAN
|
||||||
| NC score | 0.272236 (rank : 22) | θ value | 0.0193708 (rank : 16) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P15408 | Gene names | FOSL2, FRA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
DBP_HUMAN
|
||||||
| NC score | 0.253295 (rank : 23) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q10586 | Gene names | DBP | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein) (TAXREB302). | |||||
|
DBP_MOUSE
|
||||||
| NC score | 0.251878 (rank : 24) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60925, Q8VCX3 | Gene names | Dbp | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein). | |||||
|
FOSL1_MOUSE
|
||||||
| NC score | 0.248668 (rank : 25) | θ value | 0.0961366 (rank : 27) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOSL1_HUMAN
|
||||||
| NC score | 0.242905 (rank : 26) | θ value | 0.0961366 (rank : 26) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOSB_HUMAN
|
||||||
| NC score | 0.235863 (rank : 27) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
FOSB_MOUSE
|
||||||
| NC score | 0.224007 (rank : 28) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
FOS_MOUSE
|
||||||
| NC score | 0.195287 (rank : 29) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P01101 | Gene names | Fos | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos). | |||||
|
FOS_HUMAN
|
||||||
| NC score | 0.194711 (rank : 30) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P01100, P18849 | Gene names | FOS, G0S7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos) (G0/G1 switch regulatory protein 7). | |||||
|
CREB5_HUMAN
|
||||||
| NC score | 0.156559 (rank : 31) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
XBP1_MOUSE
|
||||||
| NC score | 0.149050 (rank : 32) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35426, Q8VHM0, Q922G5, Q9ESS3 | Gene names | Xbp1, Treb5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5 homolog). | |||||
|
ATF7_MOUSE
|
||||||
| NC score | 0.135938 (rank : 33) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ATF7_HUMAN
|
||||||
| NC score | 0.135218 (rank : 34) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
XBP1_HUMAN
|
||||||
| NC score | 0.129398 (rank : 35) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P17861, Q8WYK6, Q969P1, Q96BD7 | Gene names | XBP1, TREB5, XBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5). | |||||
|
CREB3_MOUSE
|
||||||
| NC score | 0.113428 (rank : 36) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61817, Q99M21, Q9CVK9 | Gene names | Creb3, Lzip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Transcription factor LZIP). | |||||
|
ATF4_HUMAN
|
||||||
| NC score | 0.100799 (rank : 37) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P18848, Q9UH31 | Gene names | ATF4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (Activating transcription factor 4) (DNA-binding protein TAXREB67) (Cyclic AMP response element-binding protein 2) (CREB2). | |||||
|
ATF4_MOUSE
|
||||||
| NC score | 0.098921 (rank : 38) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q06507, Q61906 | Gene names | Atf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (C/EBP-related ATF) (C/ATF) (TAXREB67 homolog). | |||||
|
ATF2_HUMAN
|
||||||
| NC score | 0.097246 (rank : 39) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
ATF2_MOUSE
|
||||||
| NC score | 0.097199 (rank : 40) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
JUNB_MOUSE
|
||||||
| NC score | 0.093053 (rank : 41) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P09450, Q8C2G9 | Gene names | Junb, Jun-b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-B. | |||||
|
JUN_HUMAN
|
||||||
| NC score | 0.092006 (rank : 42) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05412, Q96G93 | Gene names | JUN | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (p39). | |||||
|
JUN_MOUSE
|
||||||
| NC score | 0.091889 (rank : 43) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05627 | Gene names | Jun | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (Jun A) (AH119). | |||||
|
JUNB_HUMAN
|
||||||
| NC score | 0.089031 (rank : 44) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P17275, Q96GH3 | Gene names | JUNB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-B. | |||||
|
JUND_HUMAN
|
||||||
| NC score | 0.077275 (rank : 45) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P17535 | Gene names | JUND | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
JUND_MOUSE
|
||||||
| NC score | 0.073378 (rank : 46) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15066 | Gene names | Jund, Jun-d, Jund1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
CREB3_HUMAN
|
||||||
| NC score | 0.069056 (rank : 47) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43889, O14671, O14919, Q96GK8, Q9H2W3, Q9UE77 | Gene names | CREB3, LZIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Luman protein) (Transcription factor LZIP-alpha). | |||||
|
STRAD_MOUSE
|
||||||
| NC score | 0.068388 (rank : 48) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60924 | Gene names | Stra13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-inducible E3 protein (Hematopoietic-specific protein E3). | |||||
|
GCM1_HUMAN
|
||||||
| NC score | 0.066765 (rank : 49) | θ value | 0.0961366 (rank : 28) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NP62, Q5T0X0, Q99468, Q9P1X3 | Gene names | GCM1, GCMA | |||
|
Domain Architecture |

|
|||||
| Description | Chorion-specific transcription factor GCMa (Glial cells missing homolog 1) (GCM motif protein 1) (hGCMa). | |||||
|
ATF6A_HUMAN
|
||||||
| NC score | 0.061615 (rank : 50) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P18850, O15139, Q5VW62, Q6IPB5, Q9UEC9 | Gene names | ATF6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 alpha (Activating transcription factor 6 alpha) (ATF6-alpha). | |||||
|
NGN3_MOUSE
|
||||||
| NC score | 0.060089 (rank : 51) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70661 | Gene names | Neurog3, Ath4b, Atoh5, Ngn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3 (Atonal protein homolog 5) (Helix-loop-helix protein mATH-4B) (MATH4B). | |||||
|
SFR11_HUMAN
|
||||||
| NC score | 0.059837 (rank : 52) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
NGN2_MOUSE
|
||||||
| NC score | 0.059698 (rank : 53) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70447, P70237 | Gene names | Neurog2, Ath4a, Atoh4, Ngn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 2 (Atonal protein homolog 4) (Helix-loop-helix protein mATH-4A) (MATH4A). | |||||
|
NGN3_HUMAN
|
||||||
| NC score | 0.057316 (rank : 54) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4Z2, Q9BY24 | Gene names | NEUROG3, NGN3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3. | |||||
|
KCNH4_HUMAN
|
||||||
| NC score | 0.045409 (rank : 55) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.044711 (rank : 56) | θ value | 0.0113563 (rank : 13) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.041032 (rank : 57) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
K2027_HUMAN
|
||||||
| NC score | 0.040303 (rank : 58) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
SGOL2_MOUSE
|
||||||
| NC score | 0.039979 (rank : 59) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TSY8, Q811I4, Q8C9C4, Q8CGJ4, Q9CS55 | Gene names | Sgol2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 2. | |||||
|
NGN2_HUMAN
|
||||||
| NC score | 0.039940 (rank : 60) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H2A3, Q8N416 | Gene names | NEUROG2, NGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogenin 2. | |||||
|
RU17_HUMAN
|
||||||
| NC score | 0.038276 (rank : 61) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P08621, P78493, P78494, Q15364, Q15686, Q15687, Q15689, Q99377, Q9UE45, Q9UE46, Q9UE47, Q9UE48, Q9UFQ6 | Gene names | SNRP70, RPU1, U1AP1 | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 snRNP 70 kDa) (snRNP70) (U1-70K). | |||||
|
SGOL2_HUMAN
|
||||||
| NC score | 0.034951 (rank : 62) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q562F6, Q53RR9, Q53T20, Q86XY4, Q8IWK2, Q8IZK1, Q8N1Q5, Q96LQ3 | Gene names | SGOL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 2 (Tripin). | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.034666 (rank : 63) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
RU17_MOUSE
|
||||||
| NC score | 0.033738 (rank : 64) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62376, Q3UIW4 | Gene names | Snrp70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 SNRNP 70 kDa) (snRNP70). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.033532 (rank : 65) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
K2C8_HUMAN
|
||||||
| NC score | 0.032668 (rank : 66) | θ value | 0.0330416 (rank : 22) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P05787, Q14099, Q14716, Q14717, Q53GJ0, Q6DHW5, Q6GMY0, Q96J60 | Gene names | KRT8, CYK8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.031335 (rank : 67) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
K2C8_MOUSE
|
||||||
| NC score | 0.031120 (rank : 68) | θ value | 0.0431538 (rank : 25) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P11679, Q3KQK5, Q3TGI1, Q3TJE1, Q3TKY7, Q61463, Q61518, Q61519 | Gene names | Krt8, Krt2-8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8) (Cytokeratin endo A). | |||||
|
CHMP6_MOUSE
|
||||||
| NC score | 0.031081 (rank : 69) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P0C0A3 | Gene names | Chmp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 6 (Chromatin-modifying protein 6). | |||||
|
ZN318_HUMAN
|
||||||
| NC score | 0.030300 (rank : 70) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
CHMP6_HUMAN
|
||||||
| NC score | 0.030292 (rank : 71) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96FZ7, Q53FU4, Q9HAE8 | Gene names | CHMP6, VPS20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 6 (Chromatin-modifying protein 6) (Vacuolar protein sorting-associated protein 20) (hVps20). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.030192 (rank : 72) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
AZI1_MOUSE
|
||||||
| NC score | 0.029687 (rank : 73) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.029375 (rank : 74) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
NTBP1_MOUSE
|
||||||
| NC score | 0.028547 (rank : 75) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BRM2, Q8CAA8 | Gene names | Scyl1bp1, Ntklbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (mNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
CRSP7_HUMAN
|
||||||
| NC score | 0.027226 (rank : 76) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
TCF7_HUMAN
|
||||||
| NC score | 0.027172 (rank : 77) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P36402, Q9UKI4 | Gene names | TCF7, TCF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.027111 (rank : 78) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
IFT74_HUMAN
|
||||||
| NC score | 0.026937 (rank : 79) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
USH1C_MOUSE
|
||||||
| NC score | 0.026544 (rank : 80) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.025770 (rank : 81) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
AMOL2_HUMAN
|
||||||
| NC score | 0.025080 (rank : 82) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y2J4, Q53EP1, Q96F99, Q9UKB4 | Gene names | AMOTL2, KIAA0989 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2 (Leman coiled-coil protein) (LCCP). | |||||
|
PER1_MOUSE
|
||||||
| NC score | 0.024734 (rank : 83) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.024721 (rank : 84) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.024556 (rank : 85) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.024194 (rank : 86) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MAFG_MOUSE
|
||||||
| NC score | 0.022532 (rank : 87) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O54790 | Gene names | Mafg | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafG (V-maf musculoaponeurotic fibrosarcoma oncogene homolog G). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.021813 (rank : 88) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.021551 (rank : 89) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
SEN54_HUMAN
|
||||||
| NC score | 0.021002 (rank : 90) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z6J9, Q86WV3, Q86XE4, Q8N9H2 | Gene names | TSEN54, SEN54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen54 (tRNA-intron endonuclease Sen54) (HsSen54). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.020795 (rank : 91) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.020039 (rank : 92) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.019215 (rank : 93) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
TCF7_MOUSE
|
||||||
| NC score | 0.017962 (rank : 94) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
TJAP1_MOUSE
|
||||||
| NC score | 0.017896 (rank : 95) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DCD5, Q8CFL7, Q8R5I2 | Gene names | Tjap1, Pilt, Tjp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
PF21A_HUMAN
|
||||||
| NC score | 0.017639 (rank : 96) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
TR19L_HUMAN
|
||||||
| NC score | 0.017078 (rank : 97) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969Z4, Q86V34, Q96JU1, Q9BUX7 | Gene names | TNFRSF19L, RELT | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 19L precursor (Receptor expressed in lymphoid tissues). | |||||
|
CO4A2_HUMAN
|
||||||
| NC score | 0.016682 (rank : 98) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
MYC_MOUSE
|
||||||
| NC score | 0.016488 (rank : 99) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
RPA1_HUMAN
|
||||||
| NC score | 0.014887 (rank : 100) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95602, Q9UEH0, Q9UFT9 | Gene names | POLR1A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
SOX7_HUMAN
|
||||||
| NC score | 0.013924 (rank : 101) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.010647 (rank : 102) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.010580 (rank : 103) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
ABL1_HUMAN
|
||||||
| NC score | 0.009296 (rank : 104) | θ value | 0.125558 (rank : 29) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
TTLL4_MOUSE
|
||||||
| NC score | 0.008143 (rank : 105) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80UG8, Q5U4C4 | Gene names | Ttll4, Kiaa0173 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.008075 (rank : 106) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
DCBD1_MOUSE
|
||||||
| NC score | 0.007931 (rank : 107) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D4J3, Q8R327, Q9D696 | Gene names | Dcbld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
SMO_MOUSE
|
||||||
| NC score | 0.007143 (rank : 108) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56726 | Gene names | Smo, Smoh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smoothened homolog precursor (SMO). | |||||
|
MEIS2_MOUSE
|
||||||
| NC score | 0.006841 (rank : 109) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97367, O35676, O35677, P97403, P97404 | Gene names | Meis2, Mrg1, Stra10 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Meis2 (Meis1-related protein 1). | |||||
|
PTPRN_HUMAN
|
||||||
| NC score | 0.006316 (rank : 110) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16849, Q08319 | Gene names | PTPRN, ICA3, ICA512 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2) (Islet cell antigen 512) (ICA 512) (Islet cell autoantigen 3). | |||||
|
LRP5_MOUSE
|
||||||
| NC score | 0.006060 (rank : 111) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91VN0, O88883, Q9R208 | Gene names | Lrp5, Lrp7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor (LRP7) (Lr3). | |||||
|
PLXB2_HUMAN
|
||||||
| NC score | 0.005168 (rank : 112) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15031, Q7KZU3, Q9BSU7 | Gene names | PLXNB2, KIAA0315 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B2 precursor (MM1). | |||||
|
HXD1_MOUSE
|
||||||
| NC score | 0.004633 (rank : 113) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01822 | Gene names | Hoxd1, Hox-4.9, Hoxd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D1 (Hox-4.9). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.003084 (rank : 114) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||