Please be patient as the page loads
|
STRAD_MOUSE
|
||||||
| SwissProt Accessions | Q60924 | Gene names | Stra13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-inducible E3 protein (Hematopoietic-specific protein E3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
STRAD_MOUSE
|
||||||
| θ value | 1.63136e-134 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q60924 | Gene names | Stra13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-inducible E3 protein (Hematopoietic-specific protein E3). | |||||
|
RPTN_HUMAN
|
||||||
| θ value | 2.0648e-12 (rank : 2) | NC score | 0.243090 (rank : 3) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 3) | NC score | 0.291878 (rank : 2) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
PC11X_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 4) | NC score | 0.082165 (rank : 20) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BZA7, Q2TJH0, Q2TJH1, Q2TJH3, Q5JVZ0, Q70LR8, Q70LS7, Q70LS8, Q70LS9, Q70LT7, Q70LT8, Q70LT9, Q70LU0, Q70LU1, Q96RV4, Q96RW0, Q9BZA6, Q9H4E0, Q9P2M0, Q9P2X5 | Gene names | PCDH11X, PCDH11, PCDHX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 X-linked precursor (Protocadherin-11) (Protocadherin on the X chromosome) (PCDH-X) (Protocadherin-S). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 5) | NC score | 0.162488 (rank : 6) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
PC11Y_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 6) | NC score | 0.081855 (rank : 21) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BZA8, Q70LR6, Q70LR8, Q70LS0, Q70LS1, Q70LS2, Q70LS3, Q70LS4, Q70LS5, Q8WY34, Q9BZA9, Q9H4E1 | Gene names | PCDH11Y, PCDH11, PCDH22, PCDHY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 Y-linked precursor (Protocadherin-11) (Protocadherin- 22) (Protocadherin on the Y chromosome) (PCDH-Y) (Protocadherin prostate cancer) (Protocadherin-PC). | |||||
|
HRG_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 7) | NC score | 0.220555 (rank : 4) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04196 | Gene names | HRG | |||
|
Domain Architecture |
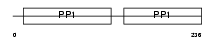
|
|||||
| Description | Histidine-rich glycoprotein precursor (Histidine-proline-rich glycoprotein) (HPRG). | |||||
|
RPTN_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 8) | NC score | 0.147367 (rank : 7) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 9) | NC score | 0.086690 (rank : 18) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
GASP2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 10) | NC score | 0.077055 (rank : 23) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96D09, Q8NAB4 | Gene names | GPRASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
SYGP1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 11) | NC score | 0.056137 (rank : 48) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
KE4_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 12) | NC score | 0.088320 (rank : 14) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q31125, Q3TVU6, Q9Z1W1 | Gene names | Slc39a7, H2-Ke4, Hke4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc transporter SLC39A7 (Solute carrier family 39 member 7) (Histidine-rich membrane protein Ke4). | |||||
|
IQEC2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.087300 (rank : 17) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 14) | NC score | 0.085545 (rank : 19) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
DAZP1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.057559 (rank : 44) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JII5 | Gene names | Dazap1 | |||
|
Domain Architecture |
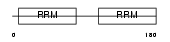
|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
ETV2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 16) | NC score | 0.051974 (rank : 54) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
REPS1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 17) | NC score | 0.061928 (rank : 39) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96D71, Q8NDR7, Q8WU62, Q9BXY9 | Gene names | REPS1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
FA47A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.203996 (rank : 5) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.068098 (rank : 35) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
DAZP1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.055093 (rank : 49) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96EP5, Q96MJ3, Q9NRR9 | Gene names | DAZAP1 | |||
|
Domain Architecture |
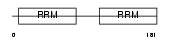
|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
BAD_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.087420 (rank : 16) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92934, O14803 | Gene names | BAD, BBC6, BCL2L8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl2 antagonist of cell death (BAD) (Bcl-2-binding component 6) (Bcl- XL/Bcl-2-associated death promoter) (Bcl-2-like 8 protein). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.043854 (rank : 68) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
CEBPA_MOUSE
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.066452 (rank : 38) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53566, Q91XB6 | Gene names | Cebpa | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
VGLL2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.072545 (rank : 27) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGW8, Q8BWB4 | Gene names | Vgll2, Vgl2, Vito1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor vestigial-like protein 2 (Vgl-2) (Protein VITO1). | |||||
|
WASL_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.116293 (rank : 9) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
CK021_HUMAN
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.108648 (rank : 12) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P2W6 | Gene names | C11orf21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf21. | |||||
|
INVS_MOUSE
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.031512 (rank : 72) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.072499 (rank : 28) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
WASL_MOUSE
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.112917 (rank : 10) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.048342 (rank : 65) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.048465 (rank : 64) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
CEBPA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.068388 (rank : 34) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49715, P78319 | Gene names | CEBPA | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
MK04_HUMAN
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.005907 (rank : 84) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31152 | Gene names | MAPK4, ERK4, PRKM4 | |||
|
Domain Architecture |
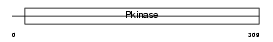
|
|||||
| Description | Mitogen-activated protein kinase 4 (EC 2.7.11.24) (Extracellular signal-regulated kinase 4) (ERK-4) (MAP kinase isoform p63) (p63- MAPK). | |||||
|
SF01_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.074005 (rank : 26) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
TAOK1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.003448 (rank : 86) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1734 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7L7X3, Q96L75, Q9H2K7, Q9H7S5, Q9P2I6 | Gene names | TAOK1, KIAA1361, MAP3K16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1) (STE20-like kinase PSK2) (Kinase from chicken homolog B) (hKFC-B). | |||||
|
FIZ1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.002741 (rank : 87) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96SL8, Q6ZMJ7 | Gene names | FIZ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flt3-interacting zinc finger protein 1. | |||||
|
INVS_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.028225 (rank : 75) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.045858 (rank : 66) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
SF01_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.069229 (rank : 31) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64213, O08817, P70167, Q61454, Q921Z4 | Gene names | Sf1, Zfm1, Zfp162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (mZFM) (Zinc finger gene in MEN1 locus) (Mammalian branch point- binding protein mBBP) (BBP) (CW17). | |||||
|
ZN302_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.001769 (rank : 88) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NR11, Q9BZD8, Q9P0J4 | Gene names | ZNF302, ZNF135L, ZNF140L | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 302 (ZNF135-like) (ZNF140-like). | |||||
|
BAG4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.045300 (rank : 67) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
CREB5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.034600 (rank : 69) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
HXB3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.021388 (rank : 78) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P14651, O95615, P17484 | Gene names | HOXB3, HOX2G | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2G) (Hox-2.7). | |||||
|
HNRPL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.030106 (rank : 73) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P14866, Q9H3P3 | Gene names | HNRPL | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L (hnRNP L). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.069087 (rank : 33) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ARSG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.014194 (rank : 82) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3TYD4, Q5XFU5, Q69ZT6, Q8CHS3, Q8VBZ5, Q9D3B4 | Gene names | Arsg, Kiaa1001 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylsulfatase G precursor (EC 3.1.6.-) (ASG). | |||||
|
ETV2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.033950 (rank : 70) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
FOXGB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.007935 (rank : 83) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60987, Q80VP3 | Gene names | Foxg1b, Fkhl1, Foxg1, Hfhbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein G1B (Forkhead-related protein FKHL1) (Transcription factor BF-1) (Brain factor 1) (BF1). | |||||
|
HNRPL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.028468 (rank : 74) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R081, O54789, Q8K0S7 | Gene names | Hnrpl | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L (hnRNP L). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.058356 (rank : 41) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
ZC3H3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.023382 (rank : 76) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ANR42_MOUSE
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.014408 (rank : 81) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3V096, Q8BX55, Q8BZT3, Q8C0T6, Q8C0X5 | Gene names | Ankrd42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
NFIX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.019413 (rank : 79) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14938, O60413, Q12859, Q13050, Q13052, Q9UPH1, Q9Y6R8 | Gene names | NFIX | |||
|
Domain Architecture |
|
|||||
| Description | Nuclear factor 1 X-type (Nuclear factor 1/X) (NF1-X) (NFI-X) (NF-I/X) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
NFIX_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.018486 (rank : 80) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70257, O08519, P70258, Q64192, Q99L78, Q9R1G2 | Gene names | Nfix | |||
|
Domain Architecture |
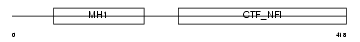
|
|||||
| Description | Nuclear factor 1 X-type (Nuclear factor 1/X) (NF1-X) (NFI-X) (NF-I/X) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
PGBM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.003855 (rank : 85) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
RBM14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.031593 (rank : 71) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96PK6, O75932, Q53GV1, Q68DQ9, Q96PK5 | Gene names | RBM14, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14) (RRM-containing coactivator activator/modulator) (Synaptotagmin-interacting protein) (SYT-interacting protein). | |||||
|
SYN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.022511 (rank : 77) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.052808 (rank : 52) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CPSF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.050403 (rank : 62) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CPSF6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.051570 (rank : 59) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.053406 (rank : 50) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
FILA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.081801 (rank : 22) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
FUS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.070896 (rank : 29) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
FUS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.067329 (rank : 36) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.069154 (rank : 32) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
HORN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.120111 (rank : 8) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
HORN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.109894 (rank : 11) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
MUC13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.102455 (rank : 13) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3R2, Q6UWD9, Q9NXT5 | Gene names | MUC13, DRCC1, RECC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-13 precursor (Down-regulated in colon cancer 1). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.061558 (rank : 40) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
MUCDL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.050437 (rank : 61) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.051649 (rank : 57) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.050113 (rank : 63) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P04554 | Gene names | PRM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protamine-2 (Protamine-P2) (Sperm histone P2) [Contains: Basic nuclear protein HPI1; Basic nuclear protein HPI2; Basic nuclear protein HPS1; Basic nuclear protein HPS2; Sperm histone HP4 (Sperm protamine P4); Sperm histone HP2 (Sperm protamine P2) (P2'); Sperm histone HP3 (Sperm protamine P3) (P2'')]. | |||||
|
PRM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.070310 (rank : 30) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P07978 | Gene names | Prm2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protamine-2 (Protamine-P2) (Sperm histone P2) [Contains: PP2-A; PP2-C; PP2-D; PP2-B (Protamine MP2)]. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.051776 (rank : 56) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
RBP56_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.075962 (rank : 24) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92804, Q92751 | Gene names | TAF15, RBP56, TAF2N | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 2N (RNA-binding protein 56) (TAFII68) (TAF(II)68). | |||||
|
S100G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.056810 (rank : 46) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29377 | Gene names | S100G, CABP9K, CALB3, S100D | |||
|
Domain Architecture |
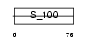
|
|||||
| Description | Protein S100-G (S100 calcium-binding protein G) (Vitamin D-dependent calcium-binding protein, intestinal) (CABP) (Calbindin D9K). | |||||
|
S10A9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.051821 (rank : 55) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31725 | Gene names | S100a9, Cagb, Mrp14 | |||
|
Domain Architecture |

|
|||||
| Description | Protein S100-A9 (S100 calcium-binding protein A9) (Calgranulin-B) (Migration inhibitory factor-related protein 14) (MRP-14) (P14) (Leukocyte L1 complex heavy chain). | |||||
|
S18L1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.057306 (rank : 45) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75177, Q5JXJ3, Q8NE69, Q9BR55, Q9H4K6 | Gene names | SS18L1, KIAA0693 | |||
|
Domain Architecture |
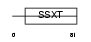
|
|||||
| Description | SS18-like protein 1 (SYT homolog 1). | |||||
|
SEMG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.051627 (rank : 58) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04279, Q6X4I9, Q6Y809, Q6Y822, Q6Y823, Q86U64, Q96QM3 | Gene names | SEMG1, SEMG | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-1 precursor (Semenogelin I) (SGI) [Contains: Alpha- inhibin-92; Alpha-inhibin-31; Seminal basic protein]. | |||||
|
SMR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.052431 (rank : 53) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
SRCH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.087716 (rank : 15) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
SSXT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.057909 (rank : 42) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15532, Q16404, Q9BXC6 | Gene names | SS18, SSXT, SYT | |||
|
Domain Architecture |
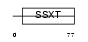
|
|||||
| Description | SSXT protein (Synovial sarcoma, translocated to X chromosome) (SYT protein). | |||||
|
SSXT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.057650 (rank : 43) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62280 | Gene names | Ss18, Ssxt, Syt | |||
|
Domain Architecture |
|
|||||
| Description | SSXT protein (SYT protein) (Synovial sarcoma-associated Ss18-alpha). | |||||
|
T2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.052944 (rank : 51) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06666 | Gene names | Srst, T2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Octapeptide-repeat protein T2. | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.050472 (rank : 60) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.075543 (rank : 25) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
WASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.067324 (rank : 37) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
ZC313_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.056332 (rank : 47) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
STRAD_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.63136e-134 (rank : 1) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q60924 | Gene names | Stra13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-inducible E3 protein (Hematopoietic-specific protein E3). | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.291878 (rank : 2) | θ value | 4.76016e-09 (rank : 3) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
RPTN_HUMAN
|
||||||
| NC score | 0.243090 (rank : 3) | θ value | 2.0648e-12 (rank : 2) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
HRG_HUMAN
|
||||||
| NC score | 0.220555 (rank : 4) | θ value | 0.000158464 (rank : 7) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04196 | Gene names | HRG | |||
|
Domain Architecture |

|
|||||
| Description | Histidine-rich glycoprotein precursor (Histidine-proline-rich glycoprotein) (HPRG). | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.203996 (rank : 5) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.162488 (rank : 6) | θ value | 8.40245e-06 (rank : 5) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.147367 (rank : 7) | θ value | 0.000158464 (rank : 8) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
HORN_HUMAN
|
||||||
| NC score | 0.120111 (rank : 8) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.116293 (rank : 9) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.112917 (rank : 10) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
HORN_MOUSE
|
||||||
| NC score | 0.109894 (rank : 11) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
CK021_HUMAN
|
||||||
| NC score | 0.108648 (rank : 12) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P2W6 | Gene names | C11orf21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf21. | |||||
|
MUC13_HUMAN
|
||||||
| NC score | 0.102455 (rank : 13) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3R2, Q6UWD9, Q9NXT5 | Gene names | MUC13, DRCC1, RECC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-13 precursor (Down-regulated in colon cancer 1). | |||||
|
KE4_MOUSE
|
||||||
| NC score | 0.088320 (rank : 14) | θ value | 0.0736092 (rank : 12) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q31125, Q3TVU6, Q9Z1W1 | Gene names | Slc39a7, H2-Ke4, Hke4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc transporter SLC39A7 (Solute carrier family 39 member 7) (Histidine-rich membrane protein Ke4). | |||||
|
SRCH_HUMAN
|
||||||
| NC score | 0.087716 (rank : 15) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
BAD_HUMAN
|
||||||
| NC score | 0.087420 (rank : 16) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92934, O14803 | Gene names | BAD, BBC6, BCL2L8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl2 antagonist of cell death (BAD) (Bcl-2-binding component 6) (Bcl- XL/Bcl-2-associated death promoter) (Bcl-2-like 8 protein). | |||||
|
IQEC2_HUMAN
|
||||||
| NC score | 0.087300 (rank : 17) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.086690 (rank : 18) | θ value | 0.00102713 (rank : 9) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
IQEC2_MOUSE
|
||||||
| NC score | 0.085545 (rank : 19) | θ value | 0.0961366 (rank : 14) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
PC11X_HUMAN
|
||||||
| NC score | 0.082165 (rank : 20) | θ value | 4.92598e-06 (rank : 4) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BZA7, Q2TJH0, Q2TJH1, Q2TJH3, Q5JVZ0, Q70LR8, Q70LS7, Q70LS8, Q70LS9, Q70LT7, Q70LT8, Q70LT9, Q70LU0, Q70LU1, Q96RV4, Q96RW0, Q9BZA6, Q9H4E0, Q9P2M0, Q9P2X5 | Gene names | PCDH11X, PCDH11, PCDHX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 X-linked precursor (Protocadherin-11) (Protocadherin on the X chromosome) (PCDH-X) (Protocadherin-S). | |||||
|
PC11Y_HUMAN
|
||||||
| NC score | 0.081855 (rank : 21) | θ value | 8.40245e-06 (rank : 6) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BZA8, Q70LR6, Q70LR8, Q70LS0, Q70LS1, Q70LS2, Q70LS3, Q70LS4, Q70LS5, Q8WY34, Q9BZA9, Q9H4E1 | Gene names | PCDH11Y, PCDH11, PCDH22, PCDHY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 Y-linked precursor (Protocadherin-11) (Protocadherin- 22) (Protocadherin on the Y chromosome) (PCDH-Y) (Protocadherin prostate cancer) (Protocadherin-PC). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.081801 (rank : 22) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
GASP2_HUMAN
|
||||||
| NC score | 0.077055 (rank : 23) | θ value | 0.0193708 (rank : 10) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96D09, Q8NAB4 | Gene names | GPRASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
RBP56_HUMAN
|
||||||
| NC score | 0.075962 (rank : 24) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92804, Q92751 | Gene names | TAF15, RBP56, TAF2N | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 2N (RNA-binding protein 56) (TAFII68) (TAF(II)68). | |||||
|
WASP_HUMAN
|
||||||
| NC score | 0.075543 (rank : 25) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
SF01_HUMAN
|
||||||
| NC score | 0.074005 (rank : 26) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
VGLL2_MOUSE
|
||||||
| NC score | 0.072545 (rank : 27) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGW8, Q8BWB4 | Gene names | Vgll2, Vgl2, Vito1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor vestigial-like protein 2 (Vgl-2) (Protein VITO1). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.072499 (rank : 28) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
FUS_HUMAN
|
||||||
| NC score | 0.070896 (rank : 29) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
PRM2_MOUSE
|
||||||
| NC score | 0.070310 (rank : 30) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P07978 | Gene names | Prm2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protamine-2 (Protamine-P2) (Sperm histone P2) [Contains: PP2-A; PP2-C; PP2-D; PP2-B (Protamine MP2)]. | |||||
|
SF01_MOUSE
|
||||||
| NC score | 0.069229 (rank : 31) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64213, O08817, P70167, Q61454, Q921Z4 | Gene names | Sf1, Zfm1, Zfp162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (mZFM) (Zinc finger gene in MEN1 locus) (Mammalian branch point- binding protein mBBP) (BBP) (CW17). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.069154 (rank : 32) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.069087 (rank : 33) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CEBPA_HUMAN
|
||||||
| NC score | 0.068388 (rank : 34) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49715, P78319 | Gene names | CEBPA | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.068098 (rank : 35) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
FUS_MOUSE
|
||||||
| NC score | 0.067329 (rank : 36) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
WASP_MOUSE
|
||||||
| NC score | 0.067324 (rank : 37) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
CEBPA_MOUSE
|
||||||
| NC score | 0.066452 (rank : 38) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53566, Q91XB6 | Gene names | Cebpa | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
REPS1_HUMAN
|
||||||
| NC score | 0.061928 (rank : 39) | θ value | 0.163984 (rank : 17) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96D71, Q8NDR7, Q8WU62, Q9BXY9 | Gene names | REPS1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.061558 (rank : 40) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.058356 (rank : 41) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
SSXT_HUMAN
|
||||||
| NC score | 0.057909 (rank : 42) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15532, Q16404, Q9BXC6 | Gene names | SS18, SSXT, SYT | |||
|
Domain Architecture |

|
|||||
| Description | SSXT protein (Synovial sarcoma, translocated to X chromosome) (SYT protein). | |||||
|
SSXT_MOUSE
|
||||||
| NC score | 0.057650 (rank : 43) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62280 | Gene names | Ss18, Ssxt, Syt | |||
|
Domain Architecture |

|
|||||
| Description | SSXT protein (SYT protein) (Synovial sarcoma-associated Ss18-alpha). | |||||
|
DAZP1_MOUSE
|
||||||
| NC score | 0.057559 (rank : 44) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JII5 | Gene names | Dazap1 | |||
|
Domain Architecture |

|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
S18L1_HUMAN
|
||||||
| NC score | 0.057306 (rank : 45) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75177, Q5JXJ3, Q8NE69, Q9BR55, Q9H4K6 | Gene names | SS18L1, KIAA0693 | |||
|
Domain Architecture |

|
|||||
| Description | SS18-like protein 1 (SYT homolog 1). | |||||
|
S100G_HUMAN
|
||||||
| NC score | 0.056810 (rank : 46) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29377 | Gene names | S100G, CABP9K, CALB3, S100D | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-G (S100 calcium-binding protein G) (Vitamin D-dependent calcium-binding protein, intestinal) (CABP) (Calbindin D9K). | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.056332 (rank : 47) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
SYGP1_HUMAN
|
||||||
| NC score | 0.056137 (rank : 48) | θ value | 0.0431538 (rank : 11) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
DAZP1_HUMAN
|
||||||
| NC score | 0.055093 (rank : 49) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96EP5, Q96MJ3, Q9NRR9 | Gene names | DAZAP1 | |||
|
Domain Architecture |

|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.053406 (rank : 50) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
T2_MOUSE
|
||||||
| NC score | 0.052944 (rank : 51) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06666 | Gene names | Srst, T2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Octapeptide-repeat protein T2. | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.052808 (rank : 52) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
SMR1_MOUSE
|
||||||
| NC score | 0.052431 (rank : 53) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
ETV2_HUMAN
|
||||||
| NC score | 0.051974 (rank : 54) | θ value | 0.125558 (rank : 16) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
S10A9_MOUSE
|
||||||
| NC score | 0.051821 (rank : 55) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31725 | Gene names | S100a9, Cagb, Mrp14 | |||
|
Domain Architecture |

|
|||||
| Description | Protein S100-A9 (S100 calcium-binding protein A9) (Calgranulin-B) (Migration inhibitory factor-related protein 14) (MRP-14) (P14) (Leukocyte L1 complex heavy chain). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.051776 (rank : 56) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.051649 (rank : 57) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
SEMG1_HUMAN
|
||||||
| NC score | 0.051627 (rank : 58) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04279, Q6X4I9, Q6Y809, Q6Y822, Q6Y823, Q86U64, Q96QM3 | Gene names | SEMG1, SEMG | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-1 precursor (Semenogelin I) (SGI) [Contains: Alpha- inhibin-92; Alpha-inhibin-31; Seminal basic protein]. | |||||
|
CPSF6_MOUSE
|
||||||
| NC score | 0.051570 (rank : 59) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.050472 (rank : 60) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
MUCDL_MOUSE
|
||||||
| NC score | 0.050437 (rank : 61) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
CPSF6_HUMAN
|
||||||
| NC score | 0.050403 (rank : 62) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
PRM2_HUMAN
|
||||||
| NC score | 0.050113 (rank : 63) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | P04554 | Gene names | PRM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protamine-2 (Protamine-P2) (Sperm histone P2) [Contains: Basic nuclear protein HPI1; Basic nuclear protein HPI2; Basic nuclear protein HPS1; Basic nuclear protein HPS2; Sperm histone HP4 (Sperm protamine P4); Sperm histone HP2 (Sperm protamine P2) (P2'); Sperm histone HP3 (Sperm protamine P3) (P2'')]. | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.048465 (rank : 64) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.048342 (rank : 65) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.045858 (rank : 66) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
BAG4_HUMAN
|
||||||
| NC score | 0.045300 (rank : 67) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.043854 (rank : 68) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
CREB5_HUMAN
|
||||||
| NC score | 0.034600 (rank : 69) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
ETV2_MOUSE
|
||||||
| NC score | 0.033950 (rank : 70) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
RBM14_HUMAN
|
||||||
| NC score | 0.031593 (rank : 71) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96PK6, O75932, Q53GV1, Q68DQ9, Q96PK5 | Gene names | RBM14, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14) (RRM-containing coactivator activator/modulator) (Synaptotagmin-interacting protein) (SYT-interacting protein). | |||||
|
INVS_MOUSE
|
||||||
| NC score | 0.031512 (rank : 72) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
HNRPL_HUMAN
|
||||||
| NC score | 0.030106 (rank : 73) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P14866, Q9H3P3 | Gene names | HNRPL | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L (hnRNP L). | |||||
|
HNRPL_MOUSE
|
||||||
| NC score | 0.028468 (rank : 74) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R081, O54789, Q8K0S7 | Gene names | Hnrpl | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L (hnRNP L). | |||||
|
INVS_HUMAN
|
||||||
| NC score | 0.028225 (rank : 75) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
ZC3H3_HUMAN
|
||||||
| NC score | 0.023382 (rank : 76) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.022511 (rank : 77) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
HXB3_HUMAN
|
||||||
| NC score | 0.021388 (rank : 78) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P14651, O95615, P17484 | Gene names | HOXB3, HOX2G | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2G) (Hox-2.7). | |||||
|
NFIX_HUMAN
|
||||||
| NC score | 0.019413 (rank : 79) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14938, O60413, Q12859, Q13050, Q13052, Q9UPH1, Q9Y6R8 | Gene names | NFIX | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 X-type (Nuclear factor 1/X) (NF1-X) (NFI-X) (NF-I/X) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
NFIX_MOUSE
|
||||||
| NC score | 0.018486 (rank : 80) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70257, O08519, P70258, Q64192, Q99L78, Q9R1G2 | Gene names | Nfix | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 X-type (Nuclear factor 1/X) (NF1-X) (NFI-X) (NF-I/X) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
ANR42_MOUSE
|
||||||
| NC score | 0.014408 (rank : 81) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3V096, Q8BX55, Q8BZT3, Q8C0T6, Q8C0X5 | Gene names | Ankrd42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
ARSG_MOUSE
|
||||||
| NC score | 0.014194 (rank : 82) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3TYD4, Q5XFU5, Q69ZT6, Q8CHS3, Q8VBZ5, Q9D3B4 | Gene names | Arsg, Kiaa1001 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylsulfatase G precursor (EC 3.1.6.-) (ASG). | |||||
|
FOXGB_MOUSE
|
||||||
| NC score | 0.007935 (rank : 83) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60987, Q80VP3 | Gene names | Foxg1b, Fkhl1, Foxg1, Hfhbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein G1B (Forkhead-related protein FKHL1) (Transcription factor BF-1) (Brain factor 1) (BF1). | |||||
|
MK04_HUMAN
|
||||||
| NC score | 0.005907 (rank : 84) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31152 | Gene names | MAPK4, ERK4, PRKM4 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 4 (EC 2.7.11.24) (Extracellular signal-regulated kinase 4) (ERK-4) (MAP kinase isoform p63) (p63- MAPK). | |||||
|
PGBM_MOUSE
|
||||||
| NC score | 0.003855 (rank : 85) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
TAOK1_HUMAN
|
||||||
| NC score | 0.003448 (rank : 86) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1734 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7L7X3, Q96L75, Q9H2K7, Q9H7S5, Q9P2I6 | Gene names | TAOK1, KIAA1361, MAP3K16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1) (STE20-like kinase PSK2) (Kinase from chicken homolog B) (hKFC-B). | |||||
|
FIZ1_HUMAN
|
||||||
| NC score | 0.002741 (rank : 87) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96SL8, Q6ZMJ7 | Gene names | FIZ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flt3-interacting zinc finger protein 1. | |||||
|
ZN302_HUMAN
|
||||||
| NC score | 0.001769 (rank : 88) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NR11, Q9BZD8, Q9P0J4 | Gene names | ZNF302, ZNF135L, ZNF140L | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 302 (ZNF135-like) (ZNF140-like). | |||||