Please be patient as the page loads
|
HNRPL_HUMAN
|
||||||
| SwissProt Accessions | P14866, Q9H3P3 | Gene names | HNRPL | |||
|
Domain Architecture |
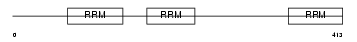
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L (hnRNP L). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HNRPL_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P14866, Q9H3P3 | Gene names | HNRPL | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L (hnRNP L). | |||||
|
HNRPL_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999731 (rank : 2) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8R081, O54789, Q8K0S7 | Gene names | Hnrpl | |||
|
Domain Architecture |
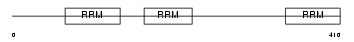
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L (hnRNP L). | |||||
|
HNRLL_HUMAN
|
||||||
| θ value | 1.57279e-145 (rank : 3) | NC score | 0.974846 (rank : 3) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WVV9, Q659B9, Q8IVH5, Q8IVH6, Q96HR5 | Gene names | HNRPLL, SRRF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L-like (Stromal RNA-regulating factor) (BLOCK24 protein). | |||||
|
HNRLL_MOUSE
|
||||||
| θ value | 2.05413e-145 (rank : 4) | NC score | 0.967674 (rank : 4) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q921F4, Q8BIP6, Q99J40 | Gene names | Hnrpll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L-like. | |||||
|
ROD1_MOUSE
|
||||||
| θ value | 7.04741e-37 (rank : 5) | NC score | 0.771164 (rank : 5) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BHD7, Q923C3 | Gene names | Rod1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of differentiation 1 (Rod1). | |||||
|
ROD1_HUMAN
|
||||||
| θ value | 3.86705e-35 (rank : 6) | NC score | 0.766507 (rank : 6) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95758, Q86YB3, Q86YH9 | Gene names | ROD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of differentiation 1 (Rod1). | |||||
|
PTBP1_HUMAN
|
||||||
| θ value | 4.43474e-23 (rank : 7) | NC score | 0.724187 (rank : 10) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26599 | Gene names | PTBP1, PTB | |||
|
Domain Architecture |

|
|||||
| Description | Polypyrimidine tract-binding protein 1 (PTB) (Heterogeneous nuclear ribonucleoprotein I) (hnRNP I) (57 kDa RNA-binding protein PPTB-1). | |||||
|
PTBP1_MOUSE
|
||||||
| θ value | 5.79196e-23 (rank : 8) | NC score | 0.739004 (rank : 7) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P17225 | Gene names | Ptbp1, Ptb | |||
|
Domain Architecture |

|
|||||
| Description | Polypyrimidine tract-binding protein 1 (PTB) (Heterogeneous nuclear ribonucleoprotein I) (hnRNP I). | |||||
|
PTBP2_MOUSE
|
||||||
| θ value | 1.29031e-22 (rank : 9) | NC score | 0.734645 (rank : 9) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91Z31, Q7TNW7, Q9CUW2, Q9QYC2, Q9R0V9 | Gene names | Ptbp2, Brptb, Nptb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypyrimidine tract-binding protein 2 (Brain-enriched polypyrimidine tract-binding protein) (Brain-enriched PTB) (RRM-type RNA-binding protein brPTB) (Neural polypyrimidine tract-binding protein). | |||||
|
PTBP2_HUMAN
|
||||||
| θ value | 1.6852e-22 (rank : 10) | NC score | 0.734917 (rank : 8) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UKA9, Q8N0Z1, Q8N160, Q8NFB0, Q8NFB1, Q969N9, Q96Q76 | Gene names | PTBP2, NPTB, PTB, PTBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypyrimidine tract-binding protein 2 (Neurally-enriched homolog of PTB) (Neural polypyrimidine tract-binding protein) (PTB-like protein). | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 11) | NC score | 0.233269 (rank : 11) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 12) | NC score | 0.224839 (rank : 12) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 13) | NC score | 0.048505 (rank : 28) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
HNRPQ_HUMAN
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.054696 (rank : 24) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60506, Q53H05, Q5TCG2, Q8IW78, Q8N599, Q96LC1, Q96LC2, Q9Y583 | Gene names | SYNCRIP, HNRPQ, NSAP1 | |||
|
Domain Architecture |
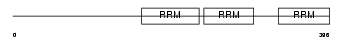
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1). | |||||
|
HNRPQ_MOUSE
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.050999 (rank : 26) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TMK9, O88991, Q80YV6, Q8BGP1, Q8C5K6, Q8CGC2, Q91ZR0, Q99KU9, Q9QYF4 | Gene names | Syncrip, Hnrpq, Nsap1, Nsap1l | |||
|
Domain Architecture |
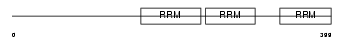
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1) (pp68). | |||||
|
RBM22_HUMAN
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.061330 (rank : 20) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NW64, O95607 | Gene names | RBM22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
RBM22_MOUSE
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.061330 (rank : 21) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BHS3, Q3TJB0, Q9CXA0 | Gene names | Rbm22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.023017 (rank : 34) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
ACF_MOUSE
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.047065 (rank : 29) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5YD48, Q5FW55, Q5YD47 | Gene names | Acf, Asp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | APOBEC1 complementation factor (APOBEC1-stimulating protein). | |||||
|
ZCRB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 20) | NC score | 0.087761 (rank : 17) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8TBF4, Q6PJX0 | Gene names | ZCRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC-type and RNA-binding motif-containing protein 1 (U11/U12 snRNP 31 kDa protein) (U11/U12-31K). | |||||
|
MK67I_HUMAN
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.042720 (rank : 30) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BYG3, Q8TB66, Q96ED4 | Gene names | MKI67IP, NIFK, NOPP34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MKI67 FHA domain-interacting nucleolar phosphoprotein (Nucleolar protein interacting with the FHA domain of pKI-67) (hNIFK) (Nucleolar phosphoprotein Nopp34). | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.106895 (rank : 14) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
ZN638_MOUSE
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.110564 (rank : 13) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.013601 (rank : 40) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PPHLN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.029117 (rank : 33) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NEY8, Q86YT2, Q8IXN3, Q8TB09, Q96NB9, Q9NXL4, Q9P0P6, Q9P0R9 | Gene names | PPHLN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Periphilin-1 (Gastric cancer antigen Ga50). | |||||
|
TIA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.022036 (rank : 36) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P31483 | Gene names | TIA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolysin TIA-1 isoform p40 (RNA-binding protein TIA-1) (p40-TIA-1) [Contains: Nucleolysin TIA-1 isoform p15 (p15-TIA-1)]. | |||||
|
STRAD_MOUSE
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.030106 (rank : 32) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60924 | Gene names | Stra13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-inducible E3 protein (Hematopoietic-specific protein E3). | |||||
|
ASTL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.008829 (rank : 43) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6HA08 | Gene names | ASTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
IQEC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.013018 (rank : 41) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.012971 (rank : 42) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
RBM23_HUMAN
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.040260 (rank : 31) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86U06, Q8ND16, Q8TB88, Q8WY40, Q9BUJ1, Q9NVV7 | Gene names | RBM23, RNPC4 | |||
|
Domain Architecture |
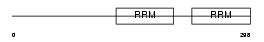
|
|||||
| Description | Probable RNA-binding protein 23 (RNA-binding motif protein 23) (RNA- binding region-containing protein 4) (Splicing factor SF2). | |||||
|
SRR35_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.022294 (rank : 35) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WXF0, Q8WW25 | Gene names | SRRP35 | |||
|
Domain Architecture |
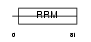
|
|||||
| Description | 35 kDa SR repressor protein (SRrp35). | |||||
|
TIA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.020037 (rank : 39) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P52912 | Gene names | Tia1, Tia | |||
|
Domain Architecture |
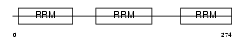
|
|||||
| Description | Nucleolysin TIA-1 (RNA-binding protein TIA-1). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.007759 (rank : 44) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
RB15B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.021576 (rank : 37) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NDT2, Q6QE19, Q9BV96 | Gene names | RBM15B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
RB15B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.021427 (rank : 38) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PHZ5, Q8C6G2 | Gene names | Rbm15b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.050290 (rank : 27) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
NONO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.054616 (rank : 25) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15233, O00201, P30807, Q12786, Q9BQC5 | Gene names | NONO, NRB54 | |||
|
Domain Architecture |
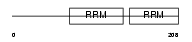
|
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein) (54 kDa nuclear RNA- and DNA-binding protein) (p54(nrb)) (p54nrb) (55 kDa nuclear protein) (NMT55) (DNA-binding p52/p100 complex, 52 kDa subunit). | |||||
|
NONO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.055217 (rank : 23) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99K48, Q63887, Q9CYQ4, Q9DBP2 | Gene names | Nono | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein). | |||||
|
RBM7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.069255 (rank : 18) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y580 | Gene names | RBM7 | |||
|
Domain Architecture |
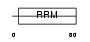
|
|||||
| Description | RNA-binding protein 7 (RNA-binding motif protein 7). | |||||
|
RBM7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.060222 (rank : 22) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CQT2, Q3UB91, Q7TQE3 | Gene names | Rbm7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 7 (RNA-binding motif protein 7). | |||||
|
RU2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.089689 (rank : 15) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08579, Q9UJD4 | Gene names | SNRPB2 | |||
|
Domain Architecture |
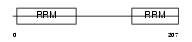
|
|||||
| Description | U2 small nuclear ribonucleoprotein B''. | |||||
|
RU2B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.088854 (rank : 16) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CQI7, Q9CW35, Q9CZ66 | Gene names | Snrpb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U2 small nuclear ribonucleoprotein B''. | |||||
|
ZCRB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.063090 (rank : 19) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CZ96, Q3TDL3 | Gene names | Zcrb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC-type and RNA-binding motif-containing protein 1 (MADP-1). | |||||
|
HNRPL_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P14866, Q9H3P3 | Gene names | HNRPL | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L (hnRNP L). | |||||
|
HNRPL_MOUSE
|
||||||
| NC score | 0.999731 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8R081, O54789, Q8K0S7 | Gene names | Hnrpl | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L (hnRNP L). | |||||
|
HNRLL_HUMAN
|
||||||
| NC score | 0.974846 (rank : 3) | θ value | 1.57279e-145 (rank : 3) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WVV9, Q659B9, Q8IVH5, Q8IVH6, Q96HR5 | Gene names | HNRPLL, SRRF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L-like (Stromal RNA-regulating factor) (BLOCK24 protein). | |||||
|
HNRLL_MOUSE
|
||||||
| NC score | 0.967674 (rank : 4) | θ value | 2.05413e-145 (rank : 4) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q921F4, Q8BIP6, Q99J40 | Gene names | Hnrpll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein L-like. | |||||
|
ROD1_MOUSE
|
||||||
| NC score | 0.771164 (rank : 5) | θ value | 7.04741e-37 (rank : 5) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BHD7, Q923C3 | Gene names | Rod1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of differentiation 1 (Rod1). | |||||
|
ROD1_HUMAN
|
||||||
| NC score | 0.766507 (rank : 6) | θ value | 3.86705e-35 (rank : 6) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95758, Q86YB3, Q86YH9 | Gene names | ROD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of differentiation 1 (Rod1). | |||||
|
PTBP1_MOUSE
|
||||||
| NC score | 0.739004 (rank : 7) | θ value | 5.79196e-23 (rank : 8) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P17225 | Gene names | Ptbp1, Ptb | |||
|
Domain Architecture |

|
|||||
| Description | Polypyrimidine tract-binding protein 1 (PTB) (Heterogeneous nuclear ribonucleoprotein I) (hnRNP I). | |||||
|
PTBP2_HUMAN
|
||||||
| NC score | 0.734917 (rank : 8) | θ value | 1.6852e-22 (rank : 10) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UKA9, Q8N0Z1, Q8N160, Q8NFB0, Q8NFB1, Q969N9, Q96Q76 | Gene names | PTBP2, NPTB, PTB, PTBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypyrimidine tract-binding protein 2 (Neurally-enriched homolog of PTB) (Neural polypyrimidine tract-binding protein) (PTB-like protein). | |||||
|
PTBP2_MOUSE
|
||||||
| NC score | 0.734645 (rank : 9) | θ value | 1.29031e-22 (rank : 9) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91Z31, Q7TNW7, Q9CUW2, Q9QYC2, Q9R0V9 | Gene names | Ptbp2, Brptb, Nptb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypyrimidine tract-binding protein 2 (Brain-enriched polypyrimidine tract-binding protein) (Brain-enriched PTB) (RRM-type RNA-binding protein brPTB) (Neural polypyrimidine tract-binding protein). | |||||
|
PTBP1_HUMAN
|
||||||
| NC score | 0.724187 (rank : 10) | θ value | 4.43474e-23 (rank : 7) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26599 | Gene names | PTBP1, PTB | |||
|
Domain Architecture |

|
|||||
| Description | Polypyrimidine tract-binding protein 1 (PTB) (Heterogeneous nuclear ribonucleoprotein I) (hnRNP I) (57 kDa RNA-binding protein PPTB-1). | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.233269 (rank : 11) | θ value | 0.000461057 (rank : 11) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.224839 (rank : 12) | θ value | 0.00509761 (rank : 12) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
ZN638_MOUSE
|
||||||
| NC score | 0.110564 (rank : 13) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.106895 (rank : 14) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
RU2B_HUMAN
|
||||||
| NC score | 0.089689 (rank : 15) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08579, Q9UJD4 | Gene names | SNRPB2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein B''. | |||||
|
RU2B_MOUSE
|
||||||
| NC score | 0.088854 (rank : 16) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CQI7, Q9CW35, Q9CZ66 | Gene names | Snrpb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U2 small nuclear ribonucleoprotein B''. | |||||
|
ZCRB1_HUMAN
|
||||||
| NC score | 0.087761 (rank : 17) | θ value | 2.36792 (rank : 20) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8TBF4, Q6PJX0 | Gene names | ZCRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC-type and RNA-binding motif-containing protein 1 (U11/U12 snRNP 31 kDa protein) (U11/U12-31K). | |||||
|
RBM7_HUMAN
|
||||||
| NC score | 0.069255 (rank : 18) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y580 | Gene names | RBM7 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 7 (RNA-binding motif protein 7). | |||||
|
ZCRB1_MOUSE
|
||||||
| NC score | 0.063090 (rank : 19) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CZ96, Q3TDL3 | Gene names | Zcrb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC-type and RNA-binding motif-containing protein 1 (MADP-1). | |||||
|
RBM22_HUMAN
|
||||||
| NC score | 0.061330 (rank : 20) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NW64, O95607 | Gene names | RBM22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
RBM22_MOUSE
|
||||||
| NC score | 0.061330 (rank : 21) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BHS3, Q3TJB0, Q9CXA0 | Gene names | Rbm22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
RBM7_MOUSE
|
||||||
| NC score | 0.060222 (rank : 22) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CQT2, Q3UB91, Q7TQE3 | Gene names | Rbm7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 7 (RNA-binding motif protein 7). | |||||
|
NONO_MOUSE
|
||||||
| NC score | 0.055217 (rank : 23) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99K48, Q63887, Q9CYQ4, Q9DBP2 | Gene names | Nono | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein). | |||||
|
HNRPQ_HUMAN
|
||||||
| NC score | 0.054696 (rank : 24) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60506, Q53H05, Q5TCG2, Q8IW78, Q8N599, Q96LC1, Q96LC2, Q9Y583 | Gene names | SYNCRIP, HNRPQ, NSAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1). | |||||
|
NONO_HUMAN
|
||||||
| NC score | 0.054616 (rank : 25) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15233, O00201, P30807, Q12786, Q9BQC5 | Gene names | NONO, NRB54 | |||
|
Domain Architecture |

|
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein) (54 kDa nuclear RNA- and DNA-binding protein) (p54(nrb)) (p54nrb) (55 kDa nuclear protein) (NMT55) (DNA-binding p52/p100 complex, 52 kDa subunit). | |||||
|
HNRPQ_MOUSE
|
||||||
| NC score | 0.050999 (rank : 26) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TMK9, O88991, Q80YV6, Q8BGP1, Q8C5K6, Q8CGC2, Q91ZR0, Q99KU9, Q9QYF4 | Gene names | Syncrip, Hnrpq, Nsap1, Nsap1l | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1) (pp68). | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.050290 (rank : 27) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.048505 (rank : 28) | θ value | 0.0330416 (rank : 13) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
ACF_MOUSE
|
||||||
| NC score | 0.047065 (rank : 29) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5YD48, Q5FW55, Q5YD47 | Gene names | Acf, Asp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | APOBEC1 complementation factor (APOBEC1-stimulating protein). | |||||
|
MK67I_HUMAN
|
||||||
| NC score | 0.042720 (rank : 30) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BYG3, Q8TB66, Q96ED4 | Gene names | MKI67IP, NIFK, NOPP34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MKI67 FHA domain-interacting nucleolar phosphoprotein (Nucleolar protein interacting with the FHA domain of pKI-67) (hNIFK) (Nucleolar phosphoprotein Nopp34). | |||||
|
RBM23_HUMAN
|
||||||
| NC score | 0.040260 (rank : 31) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86U06, Q8ND16, Q8TB88, Q8WY40, Q9BUJ1, Q9NVV7 | Gene names | RBM23, RNPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable RNA-binding protein 23 (RNA-binding motif protein 23) (RNA- binding region-containing protein 4) (Splicing factor SF2). | |||||
|
STRAD_MOUSE
|
||||||
| NC score | 0.030106 (rank : 32) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60924 | Gene names | Stra13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-inducible E3 protein (Hematopoietic-specific protein E3). | |||||
|
PPHLN_HUMAN
|
||||||
| NC score | 0.029117 (rank : 33) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NEY8, Q86YT2, Q8IXN3, Q8TB09, Q96NB9, Q9NXL4, Q9P0P6, Q9P0R9 | Gene names | PPHLN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Periphilin-1 (Gastric cancer antigen Ga50). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.023017 (rank : 34) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
SRR35_HUMAN
|
||||||
| NC score | 0.022294 (rank : 35) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WXF0, Q8WW25 | Gene names | SRRP35 | |||
|
Domain Architecture |

|
|||||
| Description | 35 kDa SR repressor protein (SRrp35). | |||||
|
TIA1_HUMAN
|
||||||
| NC score | 0.022036 (rank : 36) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P31483 | Gene names | TIA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolysin TIA-1 isoform p40 (RNA-binding protein TIA-1) (p40-TIA-1) [Contains: Nucleolysin TIA-1 isoform p15 (p15-TIA-1)]. | |||||
|
RB15B_HUMAN
|
||||||
| NC score | 0.021576 (rank : 37) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NDT2, Q6QE19, Q9BV96 | Gene names | RBM15B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
RB15B_MOUSE
|
||||||
| NC score | 0.021427 (rank : 38) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PHZ5, Q8C6G2 | Gene names | Rbm15b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
TIA1_MOUSE
|
||||||
| NC score | 0.020037 (rank : 39) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P52912 | Gene names | Tia1, Tia | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolysin TIA-1 (RNA-binding protein TIA-1). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.013601 (rank : 40) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
IQEC2_HUMAN
|
||||||
| NC score | 0.013018 (rank : 41) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_MOUSE
|
||||||
| NC score | 0.012971 (rank : 42) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
ASTL_HUMAN
|
||||||
| NC score | 0.008829 (rank : 43) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6HA08 | Gene names | ASTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.007759 (rank : 44) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||