Please be patient as the page loads
|
ZDHC5_HUMAN
|
||||||
| SwissProt Accessions | Q9C0B5, Q6ZMF0, Q8TAK8, Q9H923, Q9UFI7 | Gene names | ZDHHC5, KIAA1748, ZNF375 | |||
|
Domain Architecture |
|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5) (Zinc finger protein 375). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ZDHC5_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9C0B5, Q6ZMF0, Q8TAK8, Q9H923, Q9UFI7 | Gene names | ZDHHC5, KIAA1748, ZNF375 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5) (Zinc finger protein 375). | |||||
|
ZDHC5_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994149 (rank : 2) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8VDZ4, Q69ZB5, Q8R2X7 | Gene names | Zdhhc5, Kiaa1748 | |||
|
Domain Architecture |
|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5). | |||||
|
ZDHC8_MOUSE
|
||||||
| θ value | 5.38058e-162 (rank : 3) | NC score | 0.962503 (rank : 4) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q5Y5T5, Q7TNF7 | Gene names | Zdhhc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8). | |||||
|
ZDHC8_HUMAN
|
||||||
| θ value | 7.27209e-159 (rank : 4) | NC score | 0.966875 (rank : 3) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9ULC8, Q6ICL1, Q6ZNF5, Q7Z6L9 | Gene names | ZDHHC8, KIAA1292, ZDHHCL1, ZNF378 | |||
|
Domain Architecture |
|
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8) (Zinc finger protein 378). | |||||
|
ZDHC9_HUMAN
|
||||||
| θ value | 1.19655e-52 (rank : 5) | NC score | 0.924991 (rank : 6) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y397, Q59EK4, Q5JSW5, Q8WWS7, Q9BPY4, Q9NSP0, Q9NVL0, Q9NVR6 | Gene names | ZDHHC9, ZNF379 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC9 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 9) (DHHC-9) (DHHC9) (Zinc finger protein 379). | |||||
|
ZDHC9_MOUSE
|
||||||
| θ value | 1.19655e-52 (rank : 6) | NC score | 0.925214 (rank : 5) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P59268, Q5BL19 | Gene names | Zdhhc9 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC9 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 9) (DHHC-9) (DHHC9). | |||||
|
ZDH14_HUMAN
|
||||||
| θ value | 6.7944e-48 (rank : 7) | NC score | 0.912539 (rank : 9) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IZN3, Q5JS07, Q5JS08, Q6PHS4, Q8IZN2, Q9H7F1 | Gene names | ZDHHC14 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC14 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 14) (DHHC-14) (NEW1 domain-containing protein) (NEW1CP). | |||||
|
ZDH14_MOUSE
|
||||||
| θ value | 6.7944e-48 (rank : 8) | NC score | 0.913444 (rank : 8) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BQQ1, Q8BNR2, Q8CFN0 | Gene names | Zdhhc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC14 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 14) (DHHC-14) (NEW1 domain-containing protein) (NEW1CP). | |||||
|
ZDH18_HUMAN
|
||||||
| θ value | 4.86918e-46 (rank : 9) | NC score | 0.923289 (rank : 7) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NUE0, Q5JYH0, Q9H020 | Gene names | ZDHHC18 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC18 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 18) (DHHC-18). | |||||
|
ZDH19_HUMAN
|
||||||
| θ value | 1.85889e-29 (rank : 10) | NC score | 0.908684 (rank : 10) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WVZ1 | Gene names | ZDHHC19 | |||
|
Domain Architecture |
|
|||||
| Description | Probable palmitoyltransferase ZDHHC19 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 19) (DHHC-19). | |||||
|
ZDH19_MOUSE
|
||||||
| θ value | 2.96777e-27 (rank : 11) | NC score | 0.895076 (rank : 11) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q810M5 | Gene names | Zdhhc19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC19 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 19) (DHHC-19). | |||||
|
ZDH20_HUMAN
|
||||||
| θ value | 1.16704e-23 (rank : 12) | NC score | 0.806329 (rank : 18) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5W0Z9, Q6NVU8 | Gene names | ZDHHC20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC20 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 20) (DHHC-20). | |||||
|
ZDH15_HUMAN
|
||||||
| θ value | 5.42112e-21 (rank : 13) | NC score | 0.810370 (rank : 16) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96MV8, Q3SY30, Q6UWH3 | Gene names | ZDHHC15 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC15 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 15) (DHHC-15). | |||||
|
ZDH15_MOUSE
|
||||||
| θ value | 5.42112e-21 (rank : 14) | NC score | 0.809489 (rank : 17) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BGJ0, Q3TV63, Q8BMB7 | Gene names | Zdhhc15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Palmitoyltransferase ZDHHC15 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 15) (DHHC-15). | |||||
|
ZDH20_MOUSE
|
||||||
| θ value | 2.06002e-20 (rank : 15) | NC score | 0.799931 (rank : 22) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5Y5T1, Q3TDL1, Q8VCL6, Q9D3Q8 | Gene names | Zdhhc20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC20 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 20) (DHHC-20). | |||||
|
ZDHC2_HUMAN
|
||||||
| θ value | 2.06002e-20 (rank : 16) | NC score | 0.796331 (rank : 24) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UIJ5 | Gene names | ZDHHC2, REAM, REC, ZNF372 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC2 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 2) (DHHC-2) (Zinc finger protein 372) (Reduced expression associated with metastasis protein) (Ream) (Reduced expression in cancer protein) (Rec). | |||||
|
ZDHC2_MOUSE
|
||||||
| θ value | 2.06002e-20 (rank : 17) | NC score | 0.797106 (rank : 23) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P59267 | Gene names | Zdhhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC2 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 2) (DHHC-2). | |||||
|
ZDH17_HUMAN
|
||||||
| θ value | 2.97466e-19 (rank : 18) | NC score | 0.445935 (rank : 41) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IUH5, O75407, Q7Z2I0, Q86W89, Q86YK0, Q9P088, Q9UPZ8 | Gene names | ZDHHC17, HIP14, HIP3, KIAA0946 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14) (Huntingtin-interacting protein 3) (Huntingtin-interacting protein HYPH) (Putative NF-kappa-B-activating protein 205) (Putative MAPK- activating protein PM11). | |||||
|
ZDH17_MOUSE
|
||||||
| θ value | 3.88503e-19 (rank : 19) | NC score | 0.451166 (rank : 40) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q80TN5, Q8BJ08, Q8BYQ2, Q8BYQ6 | Gene names | Zdhhc17, Hip14, Kiaa0946 | |||
|
Domain Architecture |
|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14). | |||||
|
ZDHC3_MOUSE
|
||||||
| θ value | 1.47631e-18 (rank : 20) | NC score | 0.805591 (rank : 20) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8R173, Q6EMK3 | Gene names | Zdhhc3, Godz, Gramp1 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC3 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 3) (DHHC-3) (Golgi-specific DHHC zinc finger protein) (GABA-A receptor-associated membrane protein 1). | |||||
|
ZDHC3_HUMAN
|
||||||
| θ value | 1.24977e-17 (rank : 21) | NC score | 0.800181 (rank : 21) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NYG2, Q53A17, Q96BL0 | Gene names | ZDHHC3, ZNF373 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC3 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 3) (DHHC-3) (Zinc finger protein 373) (DHHC1 protein). | |||||
|
ZDHC4_MOUSE
|
||||||
| θ value | 8.10077e-17 (rank : 22) | NC score | 0.827442 (rank : 14) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9D6H5, Q8R5E0 | Gene names | Zdhhc4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC4 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 4) (DHHC-4). | |||||
|
ZDHC7_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 23) | NC score | 0.788102 (rank : 31) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NXF8, Q8WV42, Q9NVD8 | Gene names | ZDHHC7, ZNF370 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC7 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 7) (DHHC-7) (Zinc finger protein 370). | |||||
|
ZDHC4_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 24) | NC score | 0.825271 (rank : 15) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NPG8, Q53EV7, Q6FIB5, Q9H0R9 | Gene names | ZDHHC4, ZNF374 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC4 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 4) (DHHC-4) (Zinc finger protein 374). | |||||
|
ZDH16_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 25) | NC score | 0.806142 (rank : 19) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9ESG8, Q3TI22, Q3UA59, Q91XC5 | Gene names | Zdhhc16, Aph2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC16 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 16) (DHHC-16) (Ablphilin-2). | |||||
|
ZDHC7_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 26) | NC score | 0.788208 (rank : 30) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q91WU6, Q3TC41, Q6EMK2, Q8K1H6 | Gene names | Zdhhc7, Gramp2 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC7 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 7) (DHHC-7) (GABA-A receptor-associated membrane protein 2). | |||||
|
ZDHC6_HUMAN
|
||||||
| θ value | 5.25075e-16 (rank : 27) | NC score | 0.791848 (rank : 28) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H6R6, Q53G45, Q96IV7, Q9H605 | Gene names | ZDHHC6, ZNF376 | |||
|
Domain Architecture |
|
|||||
| Description | Probable palmitoyltransferase ZDHHC6 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 6) (DHHC-6) (Zinc finger protein 376) (Transmembrane protein H4). | |||||
|
ZDHC6_MOUSE
|
||||||
| θ value | 5.25075e-16 (rank : 28) | NC score | 0.793822 (rank : 26) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CPV7, Q3U3I8, Q3UT70, Q544H1 | Gene names | Zdhhc6 | |||
|
Domain Architecture |
|
|||||
| Description | Probable palmitoyltransferase ZDHHC6 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 6) (DHHC-6) (H4 homolog). | |||||
|
ZDH12_MOUSE
|
||||||
| θ value | 1.52774e-15 (rank : 29) | NC score | 0.851419 (rank : 12) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8VC90, Q9CZ24, Q9D0T6 | Gene names | Zdhhc12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC12 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 12) (DHHC-12). | |||||
|
ZDH12_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 30) | NC score | 0.843703 (rank : 13) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96GR4, Q5T265, Q5T267, Q5T268, Q86VT5, Q96T09 | Gene names | ZDHHC12, ZNF400 | |||
|
Domain Architecture |
|
|||||
| Description | Probable palmitoyltransferase ZDHHC12 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 12) (DHHC-12) (Zinc finger protein 400). | |||||
|
ZDH16_HUMAN
|
||||||
| θ value | 1.68911e-14 (rank : 31) | NC score | 0.796138 (rank : 25) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q969W1, Q5JTG7, Q5JTH0, Q8N4Z6, Q8WY84, Q9BSV3 | Gene names | ZDHHC16 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC16 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 16) (DHHC-16). | |||||
|
ZDH11_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 32) | NC score | 0.785740 (rank : 32) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H8X9 | Gene names | ZDHHC11, ZNF399 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC11 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 11) (DHHC-11) (Zinc finger protein 399). | |||||
|
ZDH23_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 33) | NC score | 0.769126 (rank : 35) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5Y5T3, Q3UH65, Q3UVA2 | Gene names | Zdhhc23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC23 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 23) (DHHC-23) (DHHC-containing protein 11). | |||||
|
ZDH13_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 34) | NC score | 0.434279 (rank : 42) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9CWU2, Q3UK32, Q3UKV1 | Gene names | Zdhhc13, Hip14l | |||
|
Domain Architecture |
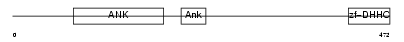
|
|||||
| Description | Probable palmitoyltransferase ZDHHC13 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 13) (DHHC-13) (Huntingtin-interacting protein 14-related protein) (HIP14-related protein). | |||||
|
ZDH21_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 35) | NC score | 0.793583 (rank : 27) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IVQ6, Q5VWG1 | Gene names | ZDHHC21 | |||
|
Domain Architecture |
|
|||||
| Description | Probable palmitoyltransferase ZDHHC21 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 21) (DHHC-21). | |||||
|
ZDH21_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 36) | NC score | 0.789510 (rank : 29) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9D270, Q6EMK1, Q80XQ3 | Gene names | Zdhhc21, Gramp3 | |||
|
Domain Architecture |
|
|||||
| Description | Probable palmitoyltransferase ZDHHC21 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 21) (DHHC-21) (GABA-A receptor-associated membrane protein 3). | |||||
|
ZDH13_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 37) | NC score | 0.428842 (rank : 43) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IUH4, Q7Z2D3, Q86VK2, Q9NV30, Q9NV99 | Gene names | ZDHHC13, HIP14L, HIP3RP | |||
|
Domain Architecture |
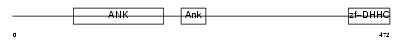
|
|||||
| Description | Probable palmitoyltransferase ZDHHC13 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 13) (DHHC-13) (Huntingtin-interacting protein 14-related protein) (HIP14-related protein) (Huntingtin- interacting protein HIP3RP) (Putative NF-kappa-B-activating protein 209) (Putative MAPK-activating protein PM03). | |||||
|
ZDHC1_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 38) | NC score | 0.737839 (rank : 38) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8WTX9, O15461 | Gene names | ZDHHC1, C16orf1, ZNF377 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC1 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 1) (DHHC-1) (Zinc finger protein 377) (DHHC- domain-containing cysteine-rich protein 1). | |||||
|
ZDH23_HUMAN
|
||||||
| θ value | 2.98157e-11 (rank : 39) | NC score | 0.749233 (rank : 36) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IYP9 | Gene names | ZDHHC23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC23 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 23) (DHHC-23). | |||||
|
ZDHC1_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 40) | NC score | 0.742391 (rank : 37) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8R0N9, Q8BJ24 | Gene names | Zdhhc1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC1 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 1) (DHHC-1). | |||||
|
ZDH24_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 41) | NC score | 0.777644 (rank : 33) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6UX98, Q6PEW7, Q9BSJ0 | Gene names | ZDHHC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC24 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 24) (DHHC-24). | |||||
|
ZDH24_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 42) | NC score | 0.770440 (rank : 34) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6IR37, Q3TAY4, Q3TDF1, Q3TF09, Q9CS66 | Gene names | Zdhhc24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC24 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 24) (DHHC-24). | |||||
|
ZDH22_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 43) | NC score | 0.712672 (rank : 39) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N966 | Gene names | ZDHHC22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative palmitoyltransferase ZDHHC22 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 22) (DHHC-22). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 44) | NC score | 0.043672 (rank : 51) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
IL16_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 45) | NC score | 0.036559 (rank : 53) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14005, Q16435, Q9UP18 | Gene names | IL16 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
TACC3_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 46) | NC score | 0.025213 (rank : 64) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
EPAS1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 47) | NC score | 0.017624 (rank : 72) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 48) | NC score | 0.050236 (rank : 48) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.163984 (rank : 49) | NC score | 0.050965 (rank : 47) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 50) | NC score | 0.029361 (rank : 55) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 51) | NC score | 0.028148 (rank : 58) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.059765 (rank : 46) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.026458 (rank : 61) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
FYB_MOUSE
|
||||||
| θ value | 0.62314 (rank : 54) | NC score | 0.035323 (rank : 54) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
SYTL2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 55) | NC score | 0.016328 (rank : 77) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.043651 (rank : 52) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
GPS2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.023036 (rank : 66) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13227 | Gene names | GPS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein pathway suppressor 2 (Protein GPS2). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.027878 (rank : 59) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
MRCKG_HUMAN
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | -0.002159 (rank : 104) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.026412 (rank : 62) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.023055 (rank : 65) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
LPIN1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.020304 (rank : 69) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZP3, Q9CQI2, Q9JLG6 | Gene names | Lpin1, Fld | |||
|
Domain Architecture |
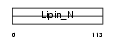
|
|||||
| Description | Lipin-1 (Fatty liver dystrophy protein). | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.006738 (rank : 90) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
ATF2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.018417 (rank : 70) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
ATF2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.017957 (rank : 71) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
CDX4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.003356 (rank : 99) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07424 | Gene names | Cdx4, Cdx-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-4 (Caudal-type homeobox protein 4). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.044131 (rank : 50) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
GAB2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.025410 (rank : 63) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.049736 (rank : 49) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
O51D1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | -0.002006 (rank : 103) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NGF3 | Gene names | OR51D1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 51D1 (Olfactory receptor OR11-14). | |||||
|
RKHD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.017301 (rank : 73) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86XN8 | Gene names | RKHD1, KIAA2031 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and KH domain-containing protein 1. | |||||
|
SEM4A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.006708 (rank : 91) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H3S1, Q8WUA9 | Gene names | SEMA4A, SEMB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
SON_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.009949 (rank : 87) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.016780 (rank : 74) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
HXB9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.004297 (rank : 97) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20615 | Gene names | Hoxb9, Hox-2.5, Hoxb-9 | |||
|
Domain Architecture |
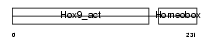
|
|||||
| Description | Homeobox protein Hox-B9 (Hox-2.5). | |||||
|
SOX7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.011098 (rank : 86) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
SVIL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.011667 (rank : 85) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
CO8A1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.015790 (rank : 78) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P27658, Q96D07 | Gene names | COL8A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VIII) chain precursor (Endothelial collagen). | |||||
|
CUTL2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.005119 (rank : 95) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
HIC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | -0.004158 (rank : 107) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R1Y5, Q9R1Y6, Q9R2B0 | Gene names | Hic1 | |||
|
Domain Architecture |
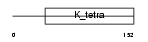
|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1). | |||||
|
HXB9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.003851 (rank : 98) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17482, Q9H1I1 | Gene names | HOXB9, HOX2E | |||
|
Domain Architecture |
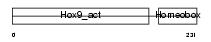
|
|||||
| Description | Homeobox protein Hox-B9 (Hox-2E) (Hox-2.5). | |||||
|
IP3KB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.020876 (rank : 68) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27987, Q5VWL9, Q5VWM0, Q96BZ2, Q96JS1, Q9UH47 | Gene names | ITPKB | |||
|
Domain Architecture |

|
|||||
| Description | Inositol-trisphosphate 3-kinase B (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase B) (IP3K B) (IP3 3-kinase B) (IP3K-B). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.028525 (rank : 56) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.013970 (rank : 82) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
SYN1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.023004 (rank : 67) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
CO4A5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.013804 (rank : 83) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
HHEX_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.002106 (rank : 100) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43120 | Gene names | Hhex, Prhx | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein PRH (Hematopoietically expressed homeobox) (Homeobox protein HEX). | |||||
|
SLIK3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.005581 (rank : 94) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94933 | Gene names | SLITRK3, KIAA0848 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SLIT and NTRK-like protein 3 precursor. | |||||
|
ATX1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.006950 (rank : 89) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
BOP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.004556 (rank : 96) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14137, Q969Z6, Q96IS8, Q9BSA7, Q9BVM0 | Gene names | BOP1, KIAA0124 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome biogenesis protein BOP1 (Block of proliferation 1 protein). | |||||
|
CO4A1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.005619 (rank : 93) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
CP135_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | -0.002516 (rank : 105) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
HSPB8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.007543 (rank : 88) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JK92 | Gene names | Hspb8, Cryac, Hsp22 | |||
|
Domain Architecture |
|
|||||
| Description | Heat-shock protein beta-8 (HspB8) (Alpha crystallin C chain) (Small stress protein-like protein HSP22). | |||||
|
K0562_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.014891 (rank : 81) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80V31, Q6ZQ95 | Gene names | Kiaa0562 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0562. | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.001805 (rank : 101) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
NEO1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | -0.001262 (rank : 102) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92859, O00340 | Gene names | NEO1, NGN | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
NU214_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.016637 (rank : 75) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
SYN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.016364 (rank : 76) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14994 | Gene names | SYN3 | |||
|
Domain Architecture |
|
|||||
| Description | Synapsin-3 (Synapsin III). | |||||
|
THUM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.015442 (rank : 80) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99J36, Q3U6Y3 | Gene names | Thumpd1 | |||
|
Domain Architecture |

|
|||||
| Description | THUMP domain-containing protein 1. | |||||
|
AKTS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.027292 (rank : 60) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96B36, Q96BI4, Q96IK7, Q96NG2, Q9BWR5 | Gene names | AKT1S1, PRAS40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich AKT1 substrate 1 (40 kDa proline-rich AKT substrate). | |||||
|
AKTS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.028222 (rank : 57) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D1F4 | Gene names | Akt1s1, Pras | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich AKT1 substrate 1 (Proline-rich AKT substrate). | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.015743 (rank : 79) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
MTR1L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | -0.002737 (rank : 106) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||
|
OCLN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.013453 (rank : 84) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61146, Q544A7 | Gene names | Ocln, Ocl | |||
|
Domain Architecture |

|
|||||
| Description | Occludin. | |||||
|
PRKDC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.006397 (rank : 92) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
ZCHC4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.082577 (rank : 45) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BKW4, Q3UTX6, Q9D2I1 | Gene names | Zcchc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 4. | |||||
|
ZCHC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.112379 (rank : 44) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H5U6, Q5IW78, Q96AN7 | Gene names | ZCCHC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 4. | |||||
|
ZDHC5_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9C0B5, Q6ZMF0, Q8TAK8, Q9H923, Q9UFI7 | Gene names | ZDHHC5, KIAA1748, ZNF375 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5) (Zinc finger protein 375). | |||||
|
ZDHC5_MOUSE
|
||||||
| NC score | 0.994149 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8VDZ4, Q69ZB5, Q8R2X7 | Gene names | Zdhhc5, Kiaa1748 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5). | |||||
|
ZDHC8_HUMAN
|
||||||
| NC score | 0.966875 (rank : 3) | θ value | 7.27209e-159 (rank : 4) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9ULC8, Q6ICL1, Q6ZNF5, Q7Z6L9 | Gene names | ZDHHC8, KIAA1292, ZDHHCL1, ZNF378 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8) (Zinc finger protein 378). | |||||
|
ZDHC8_MOUSE
|
||||||
| NC score | 0.962503 (rank : 4) | θ value | 5.38058e-162 (rank : 3) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q5Y5T5, Q7TNF7 | Gene names | Zdhhc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8). | |||||
|
ZDHC9_MOUSE
|
||||||
| NC score | 0.925214 (rank : 5) | θ value | 1.19655e-52 (rank : 6) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P59268, Q5BL19 | Gene names | Zdhhc9 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC9 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 9) (DHHC-9) (DHHC9). | |||||
|
ZDHC9_HUMAN
|
||||||
| NC score | 0.924991 (rank : 6) | θ value | 1.19655e-52 (rank : 5) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y397, Q59EK4, Q5JSW5, Q8WWS7, Q9BPY4, Q9NSP0, Q9NVL0, Q9NVR6 | Gene names | ZDHHC9, ZNF379 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC9 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 9) (DHHC-9) (DHHC9) (Zinc finger protein 379). | |||||
|
ZDH18_HUMAN
|
||||||
| NC score | 0.923289 (rank : 7) | θ value | 4.86918e-46 (rank : 9) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NUE0, Q5JYH0, Q9H020 | Gene names | ZDHHC18 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC18 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 18) (DHHC-18). | |||||
|
ZDH14_MOUSE
|
||||||
| NC score | 0.913444 (rank : 8) | θ value | 6.7944e-48 (rank : 8) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BQQ1, Q8BNR2, Q8CFN0 | Gene names | Zdhhc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC14 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 14) (DHHC-14) (NEW1 domain-containing protein) (NEW1CP). | |||||
|
ZDH14_HUMAN
|
||||||
| NC score | 0.912539 (rank : 9) | θ value | 6.7944e-48 (rank : 7) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IZN3, Q5JS07, Q5JS08, Q6PHS4, Q8IZN2, Q9H7F1 | Gene names | ZDHHC14 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC14 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 14) (DHHC-14) (NEW1 domain-containing protein) (NEW1CP). | |||||
|
ZDH19_HUMAN
|
||||||
| NC score | 0.908684 (rank : 10) | θ value | 1.85889e-29 (rank : 10) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WVZ1 | Gene names | ZDHHC19 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC19 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 19) (DHHC-19). | |||||
|
ZDH19_MOUSE
|
||||||
| NC score | 0.895076 (rank : 11) | θ value | 2.96777e-27 (rank : 11) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q810M5 | Gene names | Zdhhc19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC19 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 19) (DHHC-19). | |||||
|
ZDH12_MOUSE
|
||||||
| NC score | 0.851419 (rank : 12) | θ value | 1.52774e-15 (rank : 29) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8VC90, Q9CZ24, Q9D0T6 | Gene names | Zdhhc12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC12 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 12) (DHHC-12). | |||||
|
ZDH12_HUMAN
|
||||||
| NC score | 0.843703 (rank : 13) | θ value | 2.60593e-15 (rank : 30) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96GR4, Q5T265, Q5T267, Q5T268, Q86VT5, Q96T09 | Gene names | ZDHHC12, ZNF400 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC12 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 12) (DHHC-12) (Zinc finger protein 400). | |||||
|
ZDHC4_MOUSE
|
||||||
| NC score | 0.827442 (rank : 14) | θ value | 8.10077e-17 (rank : 22) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9D6H5, Q8R5E0 | Gene names | Zdhhc4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC4 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 4) (DHHC-4). | |||||
|
ZDHC4_HUMAN
|
||||||
| NC score | 0.825271 (rank : 15) | θ value | 1.80466e-16 (rank : 24) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NPG8, Q53EV7, Q6FIB5, Q9H0R9 | Gene names | ZDHHC4, ZNF374 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC4 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 4) (DHHC-4) (Zinc finger protein 374). | |||||
|
ZDH15_HUMAN
|
||||||
| NC score | 0.810370 (rank : 16) | θ value | 5.42112e-21 (rank : 13) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96MV8, Q3SY30, Q6UWH3 | Gene names | ZDHHC15 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC15 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 15) (DHHC-15). | |||||
|
ZDH15_MOUSE
|
||||||
| NC score | 0.809489 (rank : 17) | θ value | 5.42112e-21 (rank : 14) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BGJ0, Q3TV63, Q8BMB7 | Gene names | Zdhhc15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Palmitoyltransferase ZDHHC15 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 15) (DHHC-15). | |||||
|
ZDH20_HUMAN
|
||||||
| NC score | 0.806329 (rank : 18) | θ value | 1.16704e-23 (rank : 12) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5W0Z9, Q6NVU8 | Gene names | ZDHHC20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC20 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 20) (DHHC-20). | |||||
|
ZDH16_MOUSE
|
||||||
| NC score | 0.806142 (rank : 19) | θ value | 4.02038e-16 (rank : 25) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9ESG8, Q3TI22, Q3UA59, Q91XC5 | Gene names | Zdhhc16, Aph2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC16 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 16) (DHHC-16) (Ablphilin-2). | |||||
|
ZDHC3_MOUSE
|
||||||
| NC score | 0.805591 (rank : 20) | θ value | 1.47631e-18 (rank : 20) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8R173, Q6EMK3 | Gene names | Zdhhc3, Godz, Gramp1 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC3 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 3) (DHHC-3) (Golgi-specific DHHC zinc finger protein) (GABA-A receptor-associated membrane protein 1). | |||||
|
ZDHC3_HUMAN
|
||||||
| NC score | 0.800181 (rank : 21) | θ value | 1.24977e-17 (rank : 21) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NYG2, Q53A17, Q96BL0 | Gene names | ZDHHC3, ZNF373 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC3 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 3) (DHHC-3) (Zinc finger protein 373) (DHHC1 protein). | |||||
|
ZDH20_MOUSE
|
||||||
| NC score | 0.799931 (rank : 22) | θ value | 2.06002e-20 (rank : 15) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5Y5T1, Q3TDL1, Q8VCL6, Q9D3Q8 | Gene names | Zdhhc20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC20 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 20) (DHHC-20). | |||||
|
ZDHC2_MOUSE
|
||||||
| NC score | 0.797106 (rank : 23) | θ value | 2.06002e-20 (rank : 17) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P59267 | Gene names | Zdhhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC2 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 2) (DHHC-2). | |||||
|
ZDHC2_HUMAN
|
||||||
| NC score | 0.796331 (rank : 24) | θ value | 2.06002e-20 (rank : 16) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UIJ5 | Gene names | ZDHHC2, REAM, REC, ZNF372 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC2 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 2) (DHHC-2) (Zinc finger protein 372) (Reduced expression associated with metastasis protein) (Ream) (Reduced expression in cancer protein) (Rec). | |||||
|
ZDH16_HUMAN
|
||||||
| NC score | 0.796138 (rank : 25) | θ value | 1.68911e-14 (rank : 31) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q969W1, Q5JTG7, Q5JTH0, Q8N4Z6, Q8WY84, Q9BSV3 | Gene names | ZDHHC16 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC16 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 16) (DHHC-16). | |||||
|
ZDHC6_MOUSE
|
||||||
| NC score | 0.793822 (rank : 26) | θ value | 5.25075e-16 (rank : 28) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CPV7, Q3U3I8, Q3UT70, Q544H1 | Gene names | Zdhhc6 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC6 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 6) (DHHC-6) (H4 homolog). | |||||
|
ZDH21_HUMAN
|
||||||
| NC score | 0.793583 (rank : 27) | θ value | 3.52202e-12 (rank : 35) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IVQ6, Q5VWG1 | Gene names | ZDHHC21 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC21 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 21) (DHHC-21). | |||||
|
ZDHC6_HUMAN
|
||||||
| NC score | 0.791848 (rank : 28) | θ value | 5.25075e-16 (rank : 27) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H6R6, Q53G45, Q96IV7, Q9H605 | Gene names | ZDHHC6, ZNF376 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC6 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 6) (DHHC-6) (Zinc finger protein 376) (Transmembrane protein H4). | |||||
|
ZDH21_MOUSE
|
||||||
| NC score | 0.789510 (rank : 29) | θ value | 1.74796e-11 (rank : 36) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9D270, Q6EMK1, Q80XQ3 | Gene names | Zdhhc21, Gramp3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC21 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 21) (DHHC-21) (GABA-A receptor-associated membrane protein 3). | |||||
|
ZDHC7_MOUSE
|
||||||
| NC score | 0.788208 (rank : 30) | θ value | 4.02038e-16 (rank : 26) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q91WU6, Q3TC41, Q6EMK2, Q8K1H6 | Gene names | Zdhhc7, Gramp2 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC7 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 7) (DHHC-7) (GABA-A receptor-associated membrane protein 2). | |||||
|
ZDHC7_HUMAN
|
||||||
| NC score | 0.788102 (rank : 31) | θ value | 1.058e-16 (rank : 23) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NXF8, Q8WV42, Q9NVD8 | Gene names | ZDHHC7, ZNF370 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC7 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 7) (DHHC-7) (Zinc finger protein 370). | |||||
|
ZDH11_HUMAN
|
||||||
| NC score | 0.785740 (rank : 32) | θ value | 4.16044e-13 (rank : 32) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H8X9 | Gene names | ZDHHC11, ZNF399 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC11 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 11) (DHHC-11) (Zinc finger protein 399). | |||||
|
ZDH24_HUMAN
|
||||||
| NC score | 0.777644 (rank : 33) | θ value | 2.52405e-10 (rank : 41) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6UX98, Q6PEW7, Q9BSJ0 | Gene names | ZDHHC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC24 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 24) (DHHC-24). | |||||
|
ZDH24_MOUSE
|
||||||
| NC score | 0.770440 (rank : 34) | θ value | 2.52405e-10 (rank : 42) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6IR37, Q3TAY4, Q3TDF1, Q3TF09, Q9CS66 | Gene names | Zdhhc24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC24 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 24) (DHHC-24). | |||||
|
ZDH23_MOUSE
|
||||||
| NC score | 0.769126 (rank : 35) | θ value | 2.69671e-12 (rank : 33) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5Y5T3, Q3UH65, Q3UVA2 | Gene names | Zdhhc23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC23 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 23) (DHHC-23) (DHHC-containing protein 11). | |||||
|
ZDH23_HUMAN
|
||||||
| NC score | 0.749233 (rank : 36) | θ value | 2.98157e-11 (rank : 39) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IYP9 | Gene names | ZDHHC23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC23 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 23) (DHHC-23). | |||||
|
ZDHC1_MOUSE
|
||||||
| NC score | 0.742391 (rank : 37) | θ value | 3.89403e-11 (rank : 40) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8R0N9, Q8BJ24 | Gene names | Zdhhc1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC1 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 1) (DHHC-1). | |||||
|
ZDHC1_HUMAN
|
||||||
| NC score | 0.737839 (rank : 38) | θ value | 2.28291e-11 (rank : 38) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8WTX9, O15461 | Gene names | ZDHHC1, C16orf1, ZNF377 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC1 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 1) (DHHC-1) (Zinc finger protein 377) (DHHC- domain-containing cysteine-rich protein 1). | |||||
|
ZDH22_HUMAN
|
||||||
| NC score | 0.712672 (rank : 39) | θ value | 0.000786445 (rank : 43) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N966 | Gene names | ZDHHC22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative palmitoyltransferase ZDHHC22 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 22) (DHHC-22). | |||||
|
ZDH17_MOUSE
|
||||||
| NC score | 0.451166 (rank : 40) | θ value | 3.88503e-19 (rank : 19) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q80TN5, Q8BJ08, Q8BYQ2, Q8BYQ6 | Gene names | Zdhhc17, Hip14, Kiaa0946 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14). | |||||
|
ZDH17_HUMAN
|
||||||
| NC score | 0.445935 (rank : 41) | θ value | 2.97466e-19 (rank : 18) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IUH5, O75407, Q7Z2I0, Q86W89, Q86YK0, Q9P088, Q9UPZ8 | Gene names | ZDHHC17, HIP14, HIP3, KIAA0946 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14) (Huntingtin-interacting protein 3) (Huntingtin-interacting protein HYPH) (Putative NF-kappa-B-activating protein 205) (Putative MAPK- activating protein PM11). | |||||
|
ZDH13_MOUSE
|
||||||
| NC score | 0.434279 (rank : 42) | θ value | 3.52202e-12 (rank : 34) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9CWU2, Q3UK32, Q3UKV1 | Gene names | Zdhhc13, Hip14l | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC13 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 13) (DHHC-13) (Huntingtin-interacting protein 14-related protein) (HIP14-related protein). | |||||
|
ZDH13_HUMAN
|
||||||
| NC score | 0.428842 (rank : 43) | θ value | 2.28291e-11 (rank : 37) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IUH4, Q7Z2D3, Q86VK2, Q9NV30, Q9NV99 | Gene names | ZDHHC13, HIP14L, HIP3RP | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC13 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 13) (DHHC-13) (Huntingtin-interacting protein 14-related protein) (HIP14-related protein) (Huntingtin- interacting protein HIP3RP) (Putative NF-kappa-B-activating protein 209) (Putative MAPK-activating protein PM03). | |||||
|
ZCHC4_HUMAN
|
||||||
| NC score | 0.112379 (rank : 44) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H5U6, Q5IW78, Q96AN7 | Gene names | ZCCHC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 4. | |||||
|
ZCHC4_MOUSE
|
||||||
| NC score | 0.082577 (rank : 45) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BKW4, Q3UTX6, Q9D2I1 | Gene names | Zcchc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 4. | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.059765 (rank : 46) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.050965 (rank : 47) | θ value | 0.163984 (rank : 49) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.050236 (rank : 48) | θ value | 0.0736092 (rank : 48) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.049736 (rank : 49) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.044131 (rank : 50) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.043672 (rank : 51) | θ value | 0.00509761 (rank : 44) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.043651 (rank : 52) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
IL16_HUMAN
|
||||||
| NC score | 0.036559 (rank : 53) | θ value | 0.00665767 (rank : 45) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14005, Q16435, Q9UP18 | Gene names | IL16 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
FYB_MOUSE
|
||||||
| NC score | 0.035323 (rank : 54) | θ value | 0.62314 (rank : 54) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.029361 (rank : 55) | θ value | 0.365318 (rank : 50) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.028525 (rank : 56) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
AKTS1_MOUSE
|
||||||
| NC score | 0.028222 (rank : 57) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D1F4 | Gene names | Akt1s1, Pras | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich AKT1 substrate 1 (Proline-rich AKT substrate). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.028148 (rank : 58) | θ value | 0.47712 (rank : 51) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.027878 (rank : 59) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
AKTS1_HUMAN
|
||||||
| NC score | 0.027292 (rank : 60) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96B36, Q96BI4, Q96IK7, Q96NG2, Q9BWR5 | Gene names | AKT1S1, PRAS40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich AKT1 substrate 1 (40 kDa proline-rich AKT substrate). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.026458 (rank : 61) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.026412 (rank : 62) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
GAB2_MOUSE
|
||||||
| NC score | 0.025410 (rank : 63) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
TACC3_HUMAN
|
||||||
| NC score | 0.025213 (rank : 64) | θ value | 0.0330416 (rank : 46) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.023055 (rank : 65) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
GPS2_HUMAN
|
||||||
| NC score | 0.023036 (rank : 66) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13227 | Gene names | GPS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein pathway suppressor 2 (Protein GPS2). | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.023004 (rank : 67) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
IP3KB_HUMAN
|
||||||
| NC score | 0.020876 (rank : 68) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27987, Q5VWL9, Q5VWM0, Q96BZ2, Q96JS1, Q9UH47 | Gene names | ITPKB | |||
|
Domain Architecture |

|
|||||
| Description | Inositol-trisphosphate 3-kinase B (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase B) (IP3K B) (IP3 3-kinase B) (IP3K-B). | |||||
|
LPIN1_MOUSE
|
||||||
| NC score | 0.020304 (rank : 69) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZP3, Q9CQI2, Q9JLG6 | Gene names | Lpin1, Fld | |||
|
Domain Architecture |

|
|||||
| Description | Lipin-1 (Fatty liver dystrophy protein). | |||||
|
ATF2_HUMAN
|
||||||
| NC score | 0.018417 (rank : 70) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
ATF2_MOUSE
|
||||||
| NC score | 0.017957 (rank : 71) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
EPAS1_HUMAN
|
||||||
| NC score | 0.017624 (rank : 72) | θ value | 0.0563607 (rank : 47) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
RKHD1_HUMAN
|
||||||
| NC score | 0.017301 (rank : 73) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86XN8 | Gene names | RKHD1, KIAA2031 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and KH domain-containing protein 1. | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.016780 (rank : 74) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.016637 (rank : 75) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
SYN3_HUMAN
|
||||||
| NC score | 0.016364 (rank : 76) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14994 | Gene names | SYN3 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-3 (Synapsin III). | |||||
|
SYTL2_HUMAN
|
||||||
| NC score | 0.016328 (rank : 77) | θ value | 0.62314 (rank : 55) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
CO8A1_HUMAN
|
||||||
| NC score | 0.015790 (rank : 78) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P27658, Q96D07 | Gene names | COL8A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VIII) chain precursor (Endothelial collagen). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.015743 (rank : 79) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
THUM1_MOUSE
|
||||||
| NC score | 0.015442 (rank : 80) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99J36, Q3U6Y3 | Gene names | Thumpd1 | |||
|
Domain Architecture |

|
|||||
| Description | THUMP domain-containing protein 1. | |||||
|
K0562_MOUSE
|
||||||
| NC score | 0.014891 (rank : 81) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80V31, Q6ZQ95 | Gene names | Kiaa0562 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0562. | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.013970 (rank : 82) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
CO4A5_HUMAN
|
||||||
| NC score | 0.013804 (rank : 83) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
OCLN_MOUSE
|
||||||
| NC score | 0.013453 (rank : 84) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61146, Q544A7 | Gene names | Ocln, Ocl | |||
|
Domain Architecture |

|
|||||
| Description | Occludin. | |||||
|
SVIL_MOUSE
|
||||||
| NC score | 0.011667 (rank : 85) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
SOX7_HUMAN
|
||||||
| NC score | 0.011098 (rank : 86) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.009949 (rank : 87) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
HSPB8_MOUSE
|
||||||
| NC score | 0.007543 (rank : 88) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JK92 | Gene names | Hspb8, Cryac, Hsp22 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein beta-8 (HspB8) (Alpha crystallin C chain) (Small stress protein-like protein HSP22). | |||||
|
ATX1_MOUSE
|
||||||
| NC score | 0.006950 (rank : 89) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.006738 (rank : 90) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
SEM4A_HUMAN
|
||||||
| NC score | 0.006708 (rank : 91) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H3S1, Q8WUA9 | Gene names | SEMA4A, SEMB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
PRKDC_HUMAN
|
||||||
| NC score | 0.006397 (rank : 92) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
CO4A1_HUMAN
|
||||||
| NC score | 0.005619 (rank : 93) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
SLIK3_HUMAN
|
||||||
| NC score | 0.005581 (rank : 94) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94933 | Gene names | SLITRK3, KIAA0848 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SLIT and NTRK-like protein 3 precursor. | |||||
|
CUTL2_MOUSE
|
||||||
| NC score | 0.005119 (rank : 95) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
BOP1_HUMAN
|
||||||
| NC score | 0.004556 (rank : 96) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14137, Q969Z6, Q96IS8, Q9BSA7, Q9BVM0 | Gene names | BOP1, KIAA0124 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome biogenesis protein BOP1 (Block of proliferation 1 protein). | |||||
|
HXB9_MOUSE
|
||||||
| NC score | 0.004297 (rank : 97) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20615 | Gene names | Hoxb9, Hox-2.5, Hoxb-9 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B9 (Hox-2.5). | |||||
|
HXB9_HUMAN
|
||||||
| NC score | 0.003851 (rank : 98) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17482, Q9H1I1 | Gene names | HOXB9, HOX2E | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B9 (Hox-2E) (Hox-2.5). | |||||
|
CDX4_MOUSE
|
||||||
| NC score | 0.003356 (rank : 99) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07424 | Gene names | Cdx4, Cdx-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-4 (Caudal-type homeobox protein 4). | |||||
|
HHEX_MOUSE
|
||||||
| NC score | 0.002106 (rank : 100) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43120 | Gene names | Hhex, Prhx | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein PRH (Hematopoietically expressed homeobox) (Homeobox protein HEX). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.001805 (rank : 101) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
NEO1_HUMAN
|
||||||
| NC score | -0.001262 (rank : 102) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92859, O00340 | Gene names | NEO1, NGN | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
O51D1_HUMAN
|
||||||
| NC score | -0.002006 (rank : 103) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NGF3 | Gene names | OR51D1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 51D1 (Olfactory receptor OR11-14). | |||||
|
MRCKG_HUMAN
|
||||||
| NC score | -0.002159 (rank : 104) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
CP135_MOUSE
|
||||||
| NC score | -0.002516 (rank : 105) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
MTR1L_HUMAN
|
||||||
| NC score | -0.002737 (rank : 106) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||
|
HIC1_MOUSE
|
||||||
| NC score | -0.004158 (rank : 107) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R1Y5, Q9R1Y6, Q9R2B0 | Gene names | Hic1 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1). | |||||