Please be patient as the page loads
|
GATA6_HUMAN
|
||||||
| SwissProt Accessions | Q92908, P78327 | Gene names | GATA6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GATA6_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q92908, P78327 | Gene names | GATA6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GATA6_MOUSE
|
||||||
| θ value | 1.27474e-179 (rank : 2) | NC score | 0.992422 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61169, P97729, Q9QZK3 | Gene names | Gata6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GATA4_HUMAN
|
||||||
| θ value | 3.69378e-78 (rank : 3) | NC score | 0.971596 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P43694 | Gene names | GATA4 | |||
|
Domain Architecture |
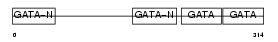
|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GATA5_MOUSE
|
||||||
| θ value | 1.5519e-76 (rank : 4) | NC score | 0.970891 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97489 | Gene names | Gata5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
GATA4_MOUSE
|
||||||
| θ value | 2.24094e-75 (rank : 5) | NC score | 0.969319 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q08369, P97491, Q9QZK4 | Gene names | Gata4, Gata-4 | |||
|
Domain Architecture |
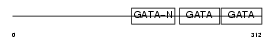
|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GATA5_HUMAN
|
||||||
| θ value | 7.97034e-73 (rank : 6) | NC score | 0.969318 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BWX5 | Gene names | GATA5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
GATA3_MOUSE
|
||||||
| θ value | 2.25659e-51 (rank : 7) | NC score | 0.932047 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P23772 | Gene names | Gata3, Gata-3 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-acting T-cell-specific transcription factor GATA-3 (GATA-binding factor 3). | |||||
|
GATA3_HUMAN
|
||||||
| θ value | 5.02714e-51 (rank : 8) | NC score | 0.930456 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P23771, Q96J16 | Gene names | GATA3 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-acting T-cell-specific transcription factor GATA-3 (GATA-binding factor 3). | |||||
|
GATA2_HUMAN
|
||||||
| θ value | 4.25575e-50 (rank : 9) | NC score | 0.924155 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P23769, Q9BUJ6 | Gene names | GATA2 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial transcription factor GATA-2 (GATA-binding protein 2). | |||||
|
GATA1_MOUSE
|
||||||
| θ value | 1.23823e-49 (rank : 10) | NC score | 0.947184 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17679 | Gene names | Gata1, Gf-1 | |||
|
Domain Architecture |

|
|||||
| Description | Erythroid transcription factor (GATA-binding factor 1) (GATA-1) (Eryf1) (GF-1) (NF-E1 DNA-binding protein). | |||||
|
GATA2_MOUSE
|
||||||
| θ value | 1.61718e-49 (rank : 11) | NC score | 0.921843 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O09100 | Gene names | Gata2 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial transcription factor GATA-2 (GATA-binding protein 2). | |||||
|
GATA1_HUMAN
|
||||||
| θ value | 1.04822e-48 (rank : 12) | NC score | 0.944740 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15976, Q96GB8 | Gene names | GATA1, ERYF1, GF1 | |||
|
Domain Architecture |

|
|||||
| Description | Erythroid transcription factor (GATA-binding factor 1) (GATA-1) (Eryf1) (GF-1) (NF-E1 DNA-binding protein). | |||||
|
TRPS1_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 13) | NC score | 0.471593 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
TRPS1_HUMAN
|
||||||
| θ value | 1.2105e-12 (rank : 14) | NC score | 0.478242 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UHF7, Q9NWE1, Q9UHH6 | Gene names | TRPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1 (Zinc finger protein GC79) (Tricho-rhino-phalangeal syndrome type I protein). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 15) | NC score | 0.009054 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.013648 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
ZN533_HUMAN
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.045859 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q569K4, Q49A04, Q6ZMZ7, Q8IY01, Q8N8H2, Q96DK4 | Gene names | ZNF533 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 533. | |||||
|
DC1L2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.024899 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PDL0 | Gene names | Dync1li2, Dncli2, Dnclic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 2 (Dynein light intermediate chain 2, cytosolic). | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.041580 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.010983 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.034961 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CO4A2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.013680 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
DZIP1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.018780 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86YF9, Q8WY45, Q8WY46, Q9UGA5, Q9Y2K0 | Gene names | DZIP1, DZIP, DZIP2, KIAA0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1/2). | |||||
|
SOX7_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.021704 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
HES6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.024148 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96HZ4, Q8N2J2, Q96T93, Q9P2S3 | Gene names | HES6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6) (C- HAIRY1). | |||||
|
CAN7_MOUSE
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.054913 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R1S8, Q9Z0P9 | Gene names | Capn7, Palbh | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.025143 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
CIC_MOUSE
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.024921 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CLSPN_MOUSE
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.021618 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
MTA2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.038849 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O94776, Q9UQB5 | Gene names | MTA2, MTA1L1, PID | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex). | |||||
|
MTA2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.038843 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R190, Q3TVT8 | Gene names | Mta2, Mta1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1). | |||||
|
NUFP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.026089 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QXX8, Q9CV69 | Gene names | Nufip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 1 (Nuclear FMRP-interacting protein 1). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.015631 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
BAP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.028846 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92560, Q6LEM0, Q7Z5E8 | Gene names | BAP1, KIAA0272 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase BAP1 (EC 3.4.19.12) (BRCA1- associated protein 1) (Cerebral protein 6). | |||||
|
HD_MOUSE
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.039921 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.007082 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
BAP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.027709 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99PU7, Q6ZQE6 | Gene names | Bap1, Kiaa0272 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase BAP1 (EC 3.4.19.12) (BRCA1- associated protein 1) (Ubiquitin C-terminal hydrolase X4) (UCH-X4). | |||||
|
HD_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.039076 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
HXB3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.005042 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P14651, O95615, P17484 | Gene names | HOXB3, HOX2G | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2G) (Hox-2.7). | |||||
|
LIMD1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.009515 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UGP4, Q9BQQ9, Q9NQ47 | Gene names | LIMD1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
CAN7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.047817 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6W3 | Gene names | CAPN7, PALBH | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
LEG4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.011145 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K419, O88353, Q91X74 | Gene names | Lgals4 | |||
|
Domain Architecture |
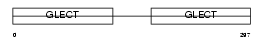
|
|||||
| Description | Galectin-4 (Lactose-binding lectin 4). | |||||
|
MYCN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.012902 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
PLXB1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.007795 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
ZCH10_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.037384 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.007206 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAF1B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.006099 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D0N7 | Gene names | Chaf1b | |||
|
Domain Architecture |
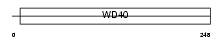
|
|||||
| Description | Chromatin assembly factor 1 subunit B (CAF-1 subunit B) (Chromatin assembly factor I p60 subunit) (CAF-I 60 kDa subunit) (CAF-Ip60). | |||||
|
CIR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.030061 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.009291 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
NEO1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.002312 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97798 | Gene names | Neo1, Ngn | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
TAU_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.021376 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
ZN533_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.035584 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BXJ8, Q8BWQ7 | Gene names | Znf533, Zfp533 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 533. | |||||
|
MUCDL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.010424 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
NOS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.008559 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29475 | Gene names | NOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, brain (EC 1.14.13.39) (NOS type I) (Neuronal NOS) (N-NOS) (nNOS) (Constitutive NOS) (NC-NOS) (bNOS). | |||||
|
OTX1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.008125 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P32242, Q53TG6 | Gene names | OTX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein OTX1. | |||||
|
SOX14_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.019569 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95416, Q3KPH7 | Gene names | SOX14, SOX28 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.018317 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P40646, Q9R1T6 | Gene names | Sox7, Sox-7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7 (mSOX7). | |||||
|
ZN11B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.000594 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 802 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06732, Q86XY8, Q8NDW3 | Gene names | ZNF11B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 11B. | |||||
|
CD008_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.015839 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78312, O43607, P78311, P78313, Q9UEG8 | Gene names | C4orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C4orf8 (Protein IT14). | |||||
|
RP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.009964 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56716 | Gene names | Rp1, Orp1, Rp1h | |||
|
Domain Architecture |
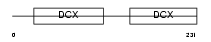
|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein homolog). | |||||
|
SIAH2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.008813 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43255, O43270 | Gene names | SIAH2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase SIAH2 (EC 6.3.2.-) (Seven in absentia homolog 2) (Siah-2) (hSiah2). | |||||
|
SP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | -0.001222 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02446, O60402 | Gene names | SP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp4 (SPR-1). | |||||
|
TNKS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.002182 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
AA2AR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | -0.001698 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29274 | Gene names | ADORA2A, ADORA2 | |||
|
Domain Architecture |
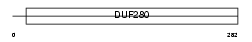
|
|||||
| Description | Adenosine A2a receptor. | |||||
|
ALEX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.012922 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
BCOR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.005086 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.015701 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
FA12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | -0.000087 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P00748, P78339 | Gene names | F12 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XII precursor (EC 3.4.21.38) (Hageman factor) (HAF) [Contains: Coagulation factor XIIa heavy chain; Beta-factor XIIa part 1; Beta-factor XIIa part 2; Coagulation factor XIIa light chain]. | |||||
|
RPA2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.010726 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70700 | Gene names | Polr1b, Rpa2, Rpo1-2 | |||
|
Domain Architecture |
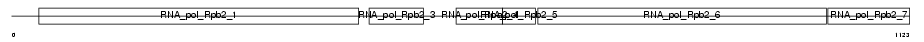
|
|||||
| Description | DNA-directed RNA polymerase I 135 kDa polypeptide (EC 2.7.7.6) (RNA polymerase I subunit 2) (RPA135). | |||||
|
SOX14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.016548 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q04892, Q9JLC9, Q9JLD0, Q9QXU4 | Gene names | Sox14, Sox-14 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX21_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.016455 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y651, P35715, Q15504 | Gene names | SOX21, SOX25, SOXA | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-21 (SOX-A). | |||||
|
SOX21_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.016448 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q811W0, Q8VH35 | Gene names | Sox21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-21. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.026343 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TNC6B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.015419 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.012436 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
CO2A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.009776 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P02458, Q12985, Q14009, Q14044, Q14046, Q14056, Q14058, Q16672, Q6LBY1, Q6LBY2, Q6LBY3, Q99227, Q9UE38, Q9UE39, Q9UE40, Q9UE41, Q9UE42, Q9UE43 | Gene names | COL2A1 | |||
|
Domain Architecture |
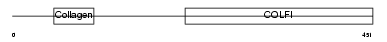
|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
CO4A2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.008580 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
FGD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.005435 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
HDAC4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.003257 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56524, Q9UND6 | Gene names | HDAC4, KIAA0288 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 4 (HD4). | |||||
|
LTK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | -0.001180 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29376 | Gene names | LTK, TYK1 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte tyrosine kinase receptor precursor (EC 2.7.10.1) (Protein tyrosine kinase 1). | |||||
|
MDFI_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.012175 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70331, P70330, P70332, Q99JM9 | Gene names | Mdfi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MyoD family inhibitor (Myogenic repressor I-mf). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.002615 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NUP98_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.010537 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P52948, Q8IUT2, Q8WYB0, Q96E54, Q9H3Q4, Q9NT02, Q9UF57, Q9UHX0, Q9Y6J4, Q9Y6J5 | Gene names | NUP98 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup98-Nup96 precursor [Contains: Nuclear pore complex protein Nup98 (Nucleoporin Nup98) (98 kDa nucleoporin); Nuclear pore complex protein Nup96 (Nucleoporin Nup96) (96 kDa nucleoporin)]. | |||||
|
OTX1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.006725 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P80205 | Gene names | Otx1, Otx-1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein OTX1. | |||||
|
RNPS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.012508 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.012508 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
SF3B2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.010651 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13435 | Gene names | SF3B2, SAP145 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 2 (Spliceosome-associated protein 145) (SAP 145) (SF3b150) (Pre-mRNA-splicing factor SF3b 145 kDa subunit). | |||||
|
UBIP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.004859 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q811S7, Q3UPR3, Q3US11, Q60786, Q8C514 | Gene names | Ubp1, Cp2b, Nf2d9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (Nuclear factor 2d9) (NF2d9). | |||||
|
GATA6_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q92908, P78327 | Gene names | GATA6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GATA6_MOUSE
|
||||||
| NC score | 0.992422 (rank : 2) | θ value | 1.27474e-179 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61169, P97729, Q9QZK3 | Gene names | Gata6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GATA4_HUMAN
|
||||||
| NC score | 0.971596 (rank : 3) | θ value | 3.69378e-78 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P43694 | Gene names | GATA4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GATA5_MOUSE
|
||||||
| NC score | 0.970891 (rank : 4) | θ value | 1.5519e-76 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97489 | Gene names | Gata5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
GATA4_MOUSE
|
||||||
| NC score | 0.969319 (rank : 5) | θ value | 2.24094e-75 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q08369, P97491, Q9QZK4 | Gene names | Gata4, Gata-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GATA5_HUMAN
|
||||||
| NC score | 0.969318 (rank : 6) | θ value | 7.97034e-73 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BWX5 | Gene names | GATA5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
GATA1_MOUSE
|
||||||
| NC score | 0.947184 (rank : 7) | θ value | 1.23823e-49 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17679 | Gene names | Gata1, Gf-1 | |||
|
Domain Architecture |

|
|||||
| Description | Erythroid transcription factor (GATA-binding factor 1) (GATA-1) (Eryf1) (GF-1) (NF-E1 DNA-binding protein). | |||||
|
GATA1_HUMAN
|
||||||
| NC score | 0.944740 (rank : 8) | θ value | 1.04822e-48 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15976, Q96GB8 | Gene names | GATA1, ERYF1, GF1 | |||
|
Domain Architecture |

|
|||||
| Description | Erythroid transcription factor (GATA-binding factor 1) (GATA-1) (Eryf1) (GF-1) (NF-E1 DNA-binding protein). | |||||
|
GATA3_MOUSE
|
||||||
| NC score | 0.932047 (rank : 9) | θ value | 2.25659e-51 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P23772 | Gene names | Gata3, Gata-3 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-acting T-cell-specific transcription factor GATA-3 (GATA-binding factor 3). | |||||
|
GATA3_HUMAN
|
||||||
| NC score | 0.930456 (rank : 10) | θ value | 5.02714e-51 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P23771, Q96J16 | Gene names | GATA3 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-acting T-cell-specific transcription factor GATA-3 (GATA-binding factor 3). | |||||
|
GATA2_HUMAN
|
||||||
| NC score | 0.924155 (rank : 11) | θ value | 4.25575e-50 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P23769, Q9BUJ6 | Gene names | GATA2 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial transcription factor GATA-2 (GATA-binding protein 2). | |||||
|
GATA2_MOUSE
|
||||||
| NC score | 0.921843 (rank : 12) | θ value | 1.61718e-49 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O09100 | Gene names | Gata2 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial transcription factor GATA-2 (GATA-binding protein 2). | |||||
|
TRPS1_HUMAN
|
||||||
| NC score | 0.478242 (rank : 13) | θ value | 1.2105e-12 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UHF7, Q9NWE1, Q9UHH6 | Gene names | TRPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1 (Zinc finger protein GC79) (Tricho-rhino-phalangeal syndrome type I protein). | |||||
|
TRPS1_MOUSE
|
||||||
| NC score | 0.471593 (rank : 14) | θ value | 2.43908e-13 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
CAN7_MOUSE
|
||||||
| NC score | 0.054913 (rank : 15) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R1S8, Q9Z0P9 | Gene names | Capn7, Palbh | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
CAN7_HUMAN
|
||||||
| NC score | 0.047817 (rank : 16) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6W3 | Gene names | CAPN7, PALBH | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
ZN533_HUMAN
|
||||||
| NC score | 0.045859 (rank : 17) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q569K4, Q49A04, Q6ZMZ7, Q8IY01, Q8N8H2, Q96DK4 | Gene names | ZNF533 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 533. | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.041580 (rank : 18) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
HD_MOUSE
|
||||||
| NC score | 0.039921 (rank : 19) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
HD_HUMAN
|
||||||
| NC score | 0.039076 (rank : 20) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
MTA2_HUMAN
|
||||||
| NC score | 0.038849 (rank : 21) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O94776, Q9UQB5 | Gene names | MTA2, MTA1L1, PID | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex). | |||||
|
MTA2_MOUSE
|
||||||
| NC score | 0.038843 (rank : 22) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R190, Q3TVT8 | Gene names | Mta2, Mta1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1). | |||||
|
ZCH10_HUMAN
|
||||||
| NC score | 0.037384 (rank : 23) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
ZN533_MOUSE
|
||||||
| NC score | 0.035584 (rank : 24) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BXJ8, Q8BWQ7 | Gene names | Znf533, Zfp533 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 533. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.034961 (rank : 25) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.030061 (rank : 26) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
BAP1_HUMAN
|
||||||
| NC score | 0.028846 (rank : 27) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92560, Q6LEM0, Q7Z5E8 | Gene names | BAP1, KIAA0272 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase BAP1 (EC 3.4.19.12) (BRCA1- associated protein 1) (Cerebral protein 6). | |||||
|
BAP1_MOUSE
|
||||||
| NC score | 0.027709 (rank : 28) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99PU7, Q6ZQE6 | Gene names | Bap1, Kiaa0272 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase BAP1 (EC 3.4.19.12) (BRCA1- associated protein 1) (Ubiquitin C-terminal hydrolase X4) (UCH-X4). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.026343 (rank : 29) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
NUFP1_MOUSE
|
||||||
| NC score | 0.026089 (rank : 30) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QXX8, Q9CV69 | Gene names | Nufip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 1 (Nuclear FMRP-interacting protein 1). | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.025143 (rank : 31) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.024921 (rank : 32) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
DC1L2_MOUSE
|
||||||
| NC score | 0.024899 (rank : 33) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PDL0 | Gene names | Dync1li2, Dncli2, Dnclic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 2 (Dynein light intermediate chain 2, cytosolic). | |||||
|
HES6_HUMAN
|
||||||
| NC score | 0.024148 (rank : 34) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96HZ4, Q8N2J2, Q96T93, Q9P2S3 | Gene names | HES6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6) (C- HAIRY1). | |||||
|
SOX7_HUMAN
|
||||||
| NC score | 0.021704 (rank : 35) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
CLSPN_MOUSE
|
||||||
| NC score | 0.021618 (rank : 36) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.021376 (rank : 37) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
SOX14_HUMAN
|
||||||
| NC score | 0.019569 (rank : 38) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95416, Q3KPH7 | Gene names | SOX14, SOX28 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
DZIP1_HUMAN
|
||||||
| NC score | 0.018780 (rank : 39) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86YF9, Q8WY45, Q8WY46, Q9UGA5, Q9Y2K0 | Gene names | DZIP1, DZIP, DZIP2, KIAA0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1/2). | |||||
|
SOX7_MOUSE
|
||||||
| NC score | 0.018317 (rank : 40) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P40646, Q9R1T6 | Gene names | Sox7, Sox-7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7 (mSOX7). | |||||
|
SOX14_MOUSE
|
||||||
| NC score | 0.016548 (rank : 41) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q04892, Q9JLC9, Q9JLD0, Q9QXU4 | Gene names | Sox14, Sox-14 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX21_HUMAN
|
||||||
| NC score | 0.016455 (rank : 42) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y651, P35715, Q15504 | Gene names | SOX21, SOX25, SOXA | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-21 (SOX-A). | |||||
|
SOX21_MOUSE
|
||||||
| NC score | 0.016448 (rank : 43) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q811W0, Q8VH35 | Gene names | Sox21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-21. | |||||
|
CD008_HUMAN
|
||||||
| NC score | 0.015839 (rank : 44) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78312, O43607, P78311, P78313, Q9UEG8 | Gene names | C4orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C4orf8 (Protein IT14). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.015701 (rank : 45) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.015631 (rank : 46) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
TNC6B_HUMAN
|
||||||
| NC score | 0.015419 (rank : 47) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
CO4A2_HUMAN
|
||||||
| NC score | 0.013680 (rank : 48) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.013648 (rank : 49) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
ALEX_MOUSE
|
||||||
| NC score | 0.012922 (rank : 50) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
MYCN_MOUSE
|
||||||
| NC score | 0.012902 (rank : 51) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
RNPS1_HUMAN
|
||||||
| NC score | 0.012508 (rank : 52) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| NC score | 0.012508 (rank : 53) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.012436 (rank : 54) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
MDFI_MOUSE
|
||||||
| NC score | 0.012175 (rank : 55) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70331, P70330, P70332, Q99JM9 | Gene names | Mdfi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MyoD family inhibitor (Myogenic repressor I-mf). | |||||
|
LEG4_MOUSE
|
||||||
| NC score | 0.011145 (rank : 56) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K419, O88353, Q91X74 | Gene names | Lgals4 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-4 (Lactose-binding lectin 4). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.010983 (rank : 57) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
RPA2_MOUSE
|
||||||
| NC score | 0.010726 (rank : 58) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70700 | Gene names | Polr1b, Rpa2, Rpo1-2 | |||
|
Domain Architecture |
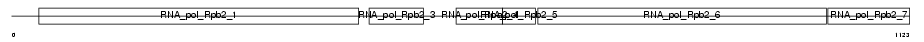
|
|||||
| Description | DNA-directed RNA polymerase I 135 kDa polypeptide (EC 2.7.7.6) (RNA polymerase I subunit 2) (RPA135). | |||||
|
SF3B2_HUMAN
|
||||||
| NC score | 0.010651 (rank : 59) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13435 | Gene names | SF3B2, SAP145 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 2 (Spliceosome-associated protein 145) (SAP 145) (SF3b150) (Pre-mRNA-splicing factor SF3b 145 kDa subunit). | |||||
|
NUP98_HUMAN
|
||||||
| NC score | 0.010537 (rank : 60) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P52948, Q8IUT2, Q8WYB0, Q96E54, Q9H3Q4, Q9NT02, Q9UF57, Q9UHX0, Q9Y6J4, Q9Y6J5 | Gene names | NUP98 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup98-Nup96 precursor [Contains: Nuclear pore complex protein Nup98 (Nucleoporin Nup98) (98 kDa nucleoporin); Nuclear pore complex protein Nup96 (Nucleoporin Nup96) (96 kDa nucleoporin)]. | |||||
|
MUCDL_MOUSE
|
||||||
| NC score | 0.010424 (rank : 61) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
RP1_MOUSE
|
||||||
| NC score | 0.009964 (rank : 62) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56716 | Gene names | Rp1, Orp1, Rp1h | |||
|
Domain Architecture |

|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein homolog). | |||||
|
CO2A1_HUMAN
|
||||||
| NC score | 0.009776 (rank : 63) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P02458, Q12985, Q14009, Q14044, Q14046, Q14056, Q14058, Q16672, Q6LBY1, Q6LBY2, Q6LBY3, Q99227, Q9UE38, Q9UE39, Q9UE40, Q9UE41, Q9UE42, Q9UE43 | Gene names | COL2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
LIMD1_HUMAN
|
||||||
| NC score | 0.009515 (rank : 64) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UGP4, Q9BQQ9, Q9NQ47 | Gene names | LIMD1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.009291 (rank : 65) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.009054 (rank : 66) | θ value | 0.0961366 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
SIAH2_HUMAN
|
||||||
| NC score | 0.008813 (rank : 67) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43255, O43270 | Gene names | SIAH2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase SIAH2 (EC 6.3.2.-) (Seven in absentia homolog 2) (Siah-2) (hSiah2). | |||||
|
CO4A2_MOUSE
|
||||||
| NC score | 0.008580 (rank : 68) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
NOS1_HUMAN
|
||||||
| NC score | 0.008559 (rank : 69) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29475 | Gene names | NOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, brain (EC 1.14.13.39) (NOS type I) (Neuronal NOS) (N-NOS) (nNOS) (Constitutive NOS) (NC-NOS) (bNOS). | |||||
|
OTX1_HUMAN
|
||||||
| NC score | 0.008125 (rank : 70) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P32242, Q53TG6 | Gene names | OTX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein OTX1. | |||||
|
PLXB1_MOUSE
|
||||||
| NC score | 0.007795 (rank : 71) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.007206 (rank : 72) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.007082 (rank : 73) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
OTX1_MOUSE
|
||||||
| NC score | 0.006725 (rank : 74) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P80205 | Gene names | Otx1, Otx-1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein OTX1. | |||||
|
CAF1B_MOUSE
|
||||||
| NC score | 0.006099 (rank : 75) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D0N7 | Gene names | Chaf1b | |||
|
Domain Architecture |

|
|||||
| Description | Chromatin assembly factor 1 subunit B (CAF-1 subunit B) (Chromatin assembly factor I p60 subunit) (CAF-I 60 kDa subunit) (CAF-Ip60). | |||||
|
FGD1_MOUSE
|
||||||
| NC score | 0.005435 (rank : 76) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
BCOR_HUMAN
|
||||||
| NC score | 0.005086 (rank : 77) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
HXB3_HUMAN
|
||||||
| NC score | 0.005042 (rank : 78) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P14651, O95615, P17484 | Gene names | HOXB3, HOX2G | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2G) (Hox-2.7). | |||||
|
UBIP1_MOUSE
|
||||||
| NC score | 0.004859 (rank : 79) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q811S7, Q3UPR3, Q3US11, Q60786, Q8C514 | Gene names | Ubp1, Cp2b, Nf2d9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (Nuclear factor 2d9) (NF2d9). | |||||
|
HDAC4_HUMAN
|
||||||
| NC score | 0.003257 (rank : 80) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56524, Q9UND6 | Gene names | HDAC4, KIAA0288 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 4 (HD4). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.002615 (rank : 81) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NEO1_MOUSE
|
||||||
| NC score | 0.002312 (rank : 82) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97798 | Gene names | Neo1, Ngn | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
TNKS1_HUMAN
|
||||||
| NC score | 0.002182 (rank : 83) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
ZN11B_HUMAN
|
||||||
| NC score | 0.000594 (rank : 84) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 802 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06732, Q86XY8, Q8NDW3 | Gene names | ZNF11B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 11B. | |||||
|
FA12_HUMAN
|
||||||
| NC score | -0.000087 (rank : 85) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P00748, P78339 | Gene names | F12 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XII precursor (EC 3.4.21.38) (Hageman factor) (HAF) [Contains: Coagulation factor XIIa heavy chain; Beta-factor XIIa part 1; Beta-factor XIIa part 2; Coagulation factor XIIa light chain]. | |||||
|
LTK_HUMAN
|
||||||
| NC score | -0.001180 (rank : 86) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29376 | Gene names | LTK, TYK1 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte tyrosine kinase receptor precursor (EC 2.7.10.1) (Protein tyrosine kinase 1). | |||||
|
SP4_HUMAN
|
||||||
| NC score | -0.001222 (rank : 87) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02446, O60402 | Gene names | SP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp4 (SPR-1). | |||||
|
AA2AR_HUMAN
|
||||||
| NC score | -0.001698 (rank : 88) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29274 | Gene names | ADORA2A, ADORA2 | |||
|
Domain Architecture |

|
|||||
| Description | Adenosine A2a receptor. | |||||