Please be patient as the page loads
|
CAN7_MOUSE
|
||||||
| SwissProt Accessions | Q9R1S8, Q9Z0P9 | Gene names | Capn7, Palbh | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CAN7_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994397 (rank : 2) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y6W3 | Gene names | CAPN7, PALBH | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
CAN7_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9R1S8, Q9Z0P9 | Gene names | Capn7, Palbh | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
CAN5_MOUSE
|
||||||
| θ value | 2.05525e-28 (rank : 3) | NC score | 0.765149 (rank : 3) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08688, Q91YU0 | Gene names | Capn5 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-5 (EC 3.4.22.-) (nCL-3). | |||||
|
CAN5_HUMAN
|
||||||
| θ value | 2.51237e-26 (rank : 4) | NC score | 0.761462 (rank : 4) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15484, O00263 | Gene names | CAPN5 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-5 (EC 3.4.22.-) (nCL-3) (htra-3). | |||||
|
CAN6_HUMAN
|
||||||
| θ value | 1.86321e-21 (rank : 5) | NC score | 0.751236 (rank : 6) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6Q1, Q9UEQ1, Q9UJA8 | Gene names | CAPN6, CALPM, CANPX | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6 (Calpamodulin) (CalpM) (Calpain-like protease X-linked). | |||||
|
CAN6_MOUSE
|
||||||
| θ value | 2.43343e-21 (rank : 6) | NC score | 0.751242 (rank : 5) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35646 | Gene names | Capn6, Capa6 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6. | |||||
|
CAN1_HUMAN
|
||||||
| θ value | 2.69047e-20 (rank : 7) | NC score | 0.669532 (rank : 16) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P07384 | Gene names | CAPN1, CANPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
CAN2_MOUSE
|
||||||
| θ value | 5.99374e-20 (rank : 8) | NC score | 0.674154 (rank : 13) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08529, O35518, O54843 | Gene names | Capn2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (80 kDa M-calpain subunit) (CALP80). | |||||
|
CAN2_HUMAN
|
||||||
| θ value | 7.82807e-20 (rank : 9) | NC score | 0.675869 (rank : 12) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17655, Q16738, Q6PJT3, Q8WU26, Q9HBB1 | Gene names | CAPN2, CANPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (Calpain large polypeptide L2). | |||||
|
CAN11_HUMAN
|
||||||
| θ value | 1.33526e-19 (rank : 10) | NC score | 0.679574 (rank : 11) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UMQ6, Q8N4R5 | Gene names | CAPN11 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-11 (EC 3.4.22.-) (Calcium-activated neutral proteinase 11) (CANP 11). | |||||
|
CAN10_HUMAN
|
||||||
| θ value | 2.27762e-19 (rank : 11) | NC score | 0.741118 (rank : 7) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HC96, Q8NCD4, Q96IG4, Q96JI2, Q9HC89, Q9HC90, Q9HC91, Q9HC92, Q9HC93, Q9HC94, Q9HC95 | Gene names | CAPN10, KIAA1845 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
CAN9_HUMAN
|
||||||
| θ value | 8.65492e-19 (rank : 12) | NC score | 0.673699 (rank : 14) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14815, Q9NS74 | Gene names | CAPN9, NCL4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4) (CG36 protein). | |||||
|
CAN1_MOUSE
|
||||||
| θ value | 1.92812e-18 (rank : 13) | NC score | 0.665310 (rank : 17) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35350, O88666 | Gene names | Capn1, Canp1, Capa1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
CAN12_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 14) | NC score | 0.698394 (rank : 9) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZSI9 | Gene names | CAPN12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain-12 (EC 3.4.22.-). | |||||
|
CAN9_MOUSE
|
||||||
| θ value | 4.29542e-18 (rank : 15) | NC score | 0.671614 (rank : 15) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D805, O35919 | Gene names | Capn9, Ncl4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4). | |||||
|
CAN10_MOUSE
|
||||||
| θ value | 2.7842e-17 (rank : 16) | NC score | 0.735274 (rank : 8) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ESK3, Q99J13, Q9WVF0 | Gene names | Capn10, Capn8 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
CAN12_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 17) | NC score | 0.687483 (rank : 10) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ER56, Q9ER53, Q9ER54, Q9ER55 | Gene names | Capn12 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-12 (EC 3.4.22.-). | |||||
|
CAN3_MOUSE
|
||||||
| θ value | 9.90251e-15 (rank : 18) | NC score | 0.646703 (rank : 19) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64691, Q9WUC5 | Gene names | Capn3 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3). | |||||
|
CAN3_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 19) | NC score | 0.647208 (rank : 18) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20807, Q9BTU4, Q9Y5S6, Q9Y5S7 | Gene names | CAPN3, CANP3, CANPL3, NCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3) (nCL-1). | |||||
|
CF103_HUMAN
|
||||||
| θ value | 1.58096e-12 (rank : 20) | NC score | 0.408132 (rank : 20) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N7X0, Q5T402 | Gene names | C6orf103 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf103. | |||||
|
VPS4B_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 21) | NC score | 0.090309 (rank : 33) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4A_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 22) | NC score | 0.087150 (rank : 34) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
VPS4B_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 23) | NC score | 0.084359 (rank : 36) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4A_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 24) | NC score | 0.085683 (rank : 35) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 25) | NC score | 0.030950 (rank : 47) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
ULK3_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 26) | NC score | 0.009195 (rank : 77) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3U3Q1, Q8K1X6, Q9DBR8 | Gene names | Ulk3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK3 (EC 2.7.11.1) (Unc-51-like kinase 3). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 0.21417 (rank : 27) | NC score | 0.018502 (rank : 62) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CE152_HUMAN
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.030226 (rank : 49) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
MYO9B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.017430 (rank : 66) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
UBP37_HUMAN
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.025413 (rank : 50) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
ARHGG_MOUSE
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.030415 (rank : 48) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3U5C8, Q501M8, Q8VCE8 | Gene names | Arhgef16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
OFD1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.040331 (rank : 45) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
GATA4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.058136 (rank : 37) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43694 | Gene names | GATA4 | |||
|
Domain Architecture |
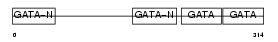
|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GATA4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.058036 (rank : 38) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08369, P97491, Q9QZK4 | Gene names | Gata4, Gata-4 | |||
|
Domain Architecture |
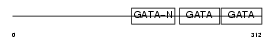
|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
SWP70_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.024015 (rank : 53) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
GATA5_MOUSE
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.057001 (rank : 39) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97489 | Gene names | Gata5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.019209 (rank : 59) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
K0146_MOUSE
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.040731 (rank : 44) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGX7, Q3TCD0, Q5U457, Q6A0B6, Q6IS60, Q811E1, Q8BWD6, Q8R305 | Gene names | Kiaa0146 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0146. | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.012104 (rank : 73) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
GATA6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.054913 (rank : 42) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92908, P78327 | Gene names | GATA6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GATA6_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.055007 (rank : 41) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61169, P97729, Q9QZK3 | Gene names | Gata6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.024683 (rank : 51) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
SPTN5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.018231 (rank : 63) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.024433 (rank : 52) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
OM34_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.021208 (rank : 55) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CYG7, Q3THD0, Q9D059 | Gene names | Tomm34 | |||
|
Domain Architecture |
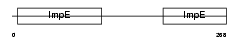
|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit). | |||||
|
OM34_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.020356 (rank : 57) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15785, Q9NTZ3 | Gene names | TOMM34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit) (hTom34). | |||||
|
RT09_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.033309 (rank : 46) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P82933 | Gene names | MRPS9, RPMS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 28S ribosomal protein S9, mitochondrial precursor (S9mt) (MRP-S9). | |||||
|
CE110_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.022332 (rank : 54) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43303, O43335, Q68DV9, Q8NE13 | Gene names | CEP110, CP110, KIAA0419 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 110 kDa (Cep110 protein). | |||||
|
GATA5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.051321 (rank : 43) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BWX5 | Gene names | GATA5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.021077 (rank : 56) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.009778 (rank : 75) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
TPSNR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.019232 (rank : 58) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VD31 | Gene names | Tapbpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tapasin-related protein precursor (TAPASIN-R) (Tapasin-like) (TAP- binding protein-related protein) (TAPBP-R) (TAP-binding protein-like). | |||||
|
ARHGG_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.017588 (rank : 64) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VV41, Q86TF0, Q99434 | Gene names | ARHGEF16, NBR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.015966 (rank : 69) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
SKIL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.018530 (rank : 61) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60665, Q60702, Q78E90, Q80VK5 | Gene names | Skil, Skir, Sno | |||
|
Domain Architecture |

|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.017564 (rank : 65) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CTGE5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.017184 (rank : 67) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 928 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R311, Q8CIE3 | Gene names | Ctage5, Mea6, Mgea6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 homolog (cTAGE-5 protein) (Meningioma-expressed antigen 6). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.015947 (rank : 70) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.015782 (rank : 71) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
PPP5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.009424 (rank : 76) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P53041, Q16722 | Gene names | PPP5C, PPP5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PP-T) (PPT). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.016388 (rank : 68) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
STX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.008665 (rank : 78) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P32856, Q86VW8 | Gene names | STX2, EPIM | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-2 (Epimorphin). | |||||
|
AR13B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.005551 (rank : 79) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3SXY8, Q504W8 | Gene names | ARL13B, ARL2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 13B (ADP-ribosylation factor-like protein 2-like 1) (ARL2-like protein 1). | |||||
|
CHM4A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.010650 (rank : 74) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BY43, Q32Q79, Q86SZ8, Q96QJ9, Q9P026 | Gene names | CHMP4A, C14orf123, SHAX2 | |||
|
Domain Architecture |
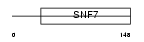
|
|||||
| Description | Charged multivesicular body protein 4a (Chromatin-modifying protein 4a) (CHMP4a) (Vacuolar protein sorting 7-1) (SNF7-1) (hSnf-1) (SNF7 homolog associated with Alix-2). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.015000 (rank : 72) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.019064 (rank : 60) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
CALL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.055446 (rank : 40) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
CPNS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.334927 (rank : 22) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P04632, Q8WTX3, Q96EW0 | Gene names | CAPNS1, CAPN4, CAPNS | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 1 (CSS1) (Calcium-dependent protease small subunit 1) (Calcium-dependent protease small subunit) (CDPS) (Calpain regulatory subunit) (Calcium-activated neutral proteinase small subunit) (CANP small subunit). | |||||
|
CPNS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.337402 (rank : 21) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88456, Q3V0X5, Q5BKQ2, Q8CEI2, Q8VEK4, Q9R1C5 | Gene names | Capns1, Capn4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 1 (CSS1) (Calcium-dependent protease small subunit 1) (Calcium-dependent protease small subunit) (CDPS) (Calpain regulatory subunit) (Calcium-activated neutral proteinase small subunit) (CANP small subunit). | |||||
|
CPNS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.332514 (rank : 23) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96L46, Q9BPV4 | Gene names | CAPNS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
CPNS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.329729 (rank : 24) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D7J7 | Gene names | Capns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
GRAN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.253983 (rank : 28) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P28676 | Gene names | GCA, GCL | |||
|
Domain Architecture |
|
|||||
| Description | Grancalcin. | |||||
|
GRAN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.259676 (rank : 25) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VC88, Q8BL53, Q8K3Z6 | Gene names | Gca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grancalcin. | |||||
|
PDCD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.195163 (rank : 31) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75340 | Gene names | PDCD6, ALG2 | |||
|
Domain Architecture |
|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2). | |||||
|
PDCD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.194257 (rank : 32) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P12815, Q545I0, Q61145 | Gene names | Pdcd6, Alg2 | |||
|
Domain Architecture |
|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2) (PMP41) (ALG-257). | |||||
|
PEF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.232863 (rank : 30) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UBV8 | Gene names | PEF1, ABP32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
PEF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.239550 (rank : 29) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BFY6, Q8VCT5, Q9CYW8, Q9D934 | Gene names | Pef1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
SORCN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.255350 (rank : 27) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P30626 | Gene names | SRI | |||
|
Domain Architecture |
|
|||||
| Description | Sorcin (22 kDa protein) (CP-22) (V19). | |||||
|
SORCN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.257521 (rank : 26) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P069, Q3UKC5, Q9CR38, Q9D7V8 | Gene names | Sri | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorcin. | |||||
|
CAN7_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9R1S8, Q9Z0P9 | Gene names | Capn7, Palbh | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
CAN7_HUMAN
|
||||||
| NC score | 0.994397 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y6W3 | Gene names | CAPN7, PALBH | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
CAN5_MOUSE
|
||||||
| NC score | 0.765149 (rank : 3) | θ value | 2.05525e-28 (rank : 3) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08688, Q91YU0 | Gene names | Capn5 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-5 (EC 3.4.22.-) (nCL-3). | |||||
|
CAN5_HUMAN
|
||||||
| NC score | 0.761462 (rank : 4) | θ value | 2.51237e-26 (rank : 4) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15484, O00263 | Gene names | CAPN5 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-5 (EC 3.4.22.-) (nCL-3) (htra-3). | |||||
|
CAN6_MOUSE
|
||||||
| NC score | 0.751242 (rank : 5) | θ value | 2.43343e-21 (rank : 6) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35646 | Gene names | Capn6, Capa6 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6. | |||||
|
CAN6_HUMAN
|
||||||
| NC score | 0.751236 (rank : 6) | θ value | 1.86321e-21 (rank : 5) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6Q1, Q9UEQ1, Q9UJA8 | Gene names | CAPN6, CALPM, CANPX | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6 (Calpamodulin) (CalpM) (Calpain-like protease X-linked). | |||||
|
CAN10_HUMAN
|
||||||
| NC score | 0.741118 (rank : 7) | θ value | 2.27762e-19 (rank : 11) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HC96, Q8NCD4, Q96IG4, Q96JI2, Q9HC89, Q9HC90, Q9HC91, Q9HC92, Q9HC93, Q9HC94, Q9HC95 | Gene names | CAPN10, KIAA1845 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
CAN10_MOUSE
|
||||||
| NC score | 0.735274 (rank : 8) | θ value | 2.7842e-17 (rank : 16) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ESK3, Q99J13, Q9WVF0 | Gene names | Capn10, Capn8 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
CAN12_HUMAN
|
||||||
| NC score | 0.698394 (rank : 9) | θ value | 2.5182e-18 (rank : 14) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZSI9 | Gene names | CAPN12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain-12 (EC 3.4.22.-). | |||||
|
CAN12_MOUSE
|
||||||
| NC score | 0.687483 (rank : 10) | θ value | 3.40345e-15 (rank : 17) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ER56, Q9ER53, Q9ER54, Q9ER55 | Gene names | Capn12 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-12 (EC 3.4.22.-). | |||||
|
CAN11_HUMAN
|
||||||
| NC score | 0.679574 (rank : 11) | θ value | 1.33526e-19 (rank : 10) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UMQ6, Q8N4R5 | Gene names | CAPN11 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-11 (EC 3.4.22.-) (Calcium-activated neutral proteinase 11) (CANP 11). | |||||
|
CAN2_HUMAN
|
||||||
| NC score | 0.675869 (rank : 12) | θ value | 7.82807e-20 (rank : 9) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17655, Q16738, Q6PJT3, Q8WU26, Q9HBB1 | Gene names | CAPN2, CANPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (Calpain large polypeptide L2). | |||||
|
CAN2_MOUSE
|
||||||
| NC score | 0.674154 (rank : 13) | θ value | 5.99374e-20 (rank : 8) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08529, O35518, O54843 | Gene names | Capn2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (80 kDa M-calpain subunit) (CALP80). | |||||
|
CAN9_HUMAN
|
||||||
| NC score | 0.673699 (rank : 14) | θ value | 8.65492e-19 (rank : 12) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14815, Q9NS74 | Gene names | CAPN9, NCL4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4) (CG36 protein). | |||||
|
CAN9_MOUSE
|
||||||
| NC score | 0.671614 (rank : 15) | θ value | 4.29542e-18 (rank : 15) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D805, O35919 | Gene names | Capn9, Ncl4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4). | |||||
|
CAN1_HUMAN
|
||||||
| NC score | 0.669532 (rank : 16) | θ value | 2.69047e-20 (rank : 7) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P07384 | Gene names | CAPN1, CANPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
CAN1_MOUSE
|
||||||
| NC score | 0.665310 (rank : 17) | θ value | 1.92812e-18 (rank : 13) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35350, O88666 | Gene names | Capn1, Canp1, Capa1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
CAN3_HUMAN
|
||||||
| NC score | 0.647208 (rank : 18) | θ value | 2.88119e-14 (rank : 19) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20807, Q9BTU4, Q9Y5S6, Q9Y5S7 | Gene names | CAPN3, CANP3, CANPL3, NCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3) (nCL-1). | |||||
|
CAN3_MOUSE
|
||||||
| NC score | 0.646703 (rank : 19) | θ value | 9.90251e-15 (rank : 18) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64691, Q9WUC5 | Gene names | Capn3 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3). | |||||
|
CF103_HUMAN
|
||||||
| NC score | 0.408132 (rank : 20) | θ value | 1.58096e-12 (rank : 20) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N7X0, Q5T402 | Gene names | C6orf103 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf103. | |||||
|
CPNS1_MOUSE
|
||||||
| NC score | 0.337402 (rank : 21) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88456, Q3V0X5, Q5BKQ2, Q8CEI2, Q8VEK4, Q9R1C5 | Gene names | Capns1, Capn4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 1 (CSS1) (Calcium-dependent protease small subunit 1) (Calcium-dependent protease small subunit) (CDPS) (Calpain regulatory subunit) (Calcium-activated neutral proteinase small subunit) (CANP small subunit). | |||||
|
CPNS1_HUMAN
|
||||||
| NC score | 0.334927 (rank : 22) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P04632, Q8WTX3, Q96EW0 | Gene names | CAPNS1, CAPN4, CAPNS | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 1 (CSS1) (Calcium-dependent protease small subunit 1) (Calcium-dependent protease small subunit) (CDPS) (Calpain regulatory subunit) (Calcium-activated neutral proteinase small subunit) (CANP small subunit). | |||||
|
CPNS2_HUMAN
|
||||||
| NC score | 0.332514 (rank : 23) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96L46, Q9BPV4 | Gene names | CAPNS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
CPNS2_MOUSE
|
||||||
| NC score | 0.329729 (rank : 24) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D7J7 | Gene names | Capns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
GRAN_MOUSE
|
||||||
| NC score | 0.259676 (rank : 25) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VC88, Q8BL53, Q8K3Z6 | Gene names | Gca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grancalcin. | |||||
|
SORCN_MOUSE
|
||||||
| NC score | 0.257521 (rank : 26) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P069, Q3UKC5, Q9CR38, Q9D7V8 | Gene names | Sri | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorcin. | |||||
|
SORCN_HUMAN
|
||||||
| NC score | 0.255350 (rank : 27) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P30626 | Gene names | SRI | |||
|
Domain Architecture |

|
|||||
| Description | Sorcin (22 kDa protein) (CP-22) (V19). | |||||
|
GRAN_HUMAN
|
||||||
| NC score | 0.253983 (rank : 28) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P28676 | Gene names | GCA, GCL | |||
|
Domain Architecture |

|
|||||
| Description | Grancalcin. | |||||
|
PEF1_MOUSE
|
||||||
| NC score | 0.239550 (rank : 29) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BFY6, Q8VCT5, Q9CYW8, Q9D934 | Gene names | Pef1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
PEF1_HUMAN
|
||||||
| NC score | 0.232863 (rank : 30) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UBV8 | Gene names | PEF1, ABP32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
PDCD6_HUMAN
|
||||||
| NC score | 0.195163 (rank : 31) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75340 | Gene names | PDCD6, ALG2 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2). | |||||
|
PDCD6_MOUSE
|
||||||
| NC score | 0.194257 (rank : 32) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P12815, Q545I0, Q61145 | Gene names | Pdcd6, Alg2 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2) (PMP41) (ALG-257). | |||||
|
VPS4B_MOUSE
|
||||||
| NC score | 0.090309 (rank : 33) | θ value | 0.000270298 (rank : 21) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
VPS4A_MOUSE
|
||||||
| NC score | 0.087150 (rank : 34) | θ value | 0.00390308 (rank : 22) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
VPS4A_HUMAN
|
||||||
| NC score | 0.085683 (rank : 35) | θ value | 0.00665767 (rank : 24) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
VPS4B_HUMAN
|
||||||
| NC score | 0.084359 (rank : 36) | θ value | 0.00390308 (rank : 23) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75351, Q9GZS7 | Gene names | VPS4B, SKD1, VPS42 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
GATA4_HUMAN
|
||||||
| NC score | 0.058136 (rank : 37) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43694 | Gene names | GATA4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GATA4_MOUSE
|
||||||
| NC score | 0.058036 (rank : 38) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08369, P97491, Q9QZK4 | Gene names | Gata4, Gata-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GATA5_MOUSE
|
||||||
| NC score | 0.057001 (rank : 39) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97489 | Gene names | Gata5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
CALL5_HUMAN
|
||||||
| NC score | 0.055446 (rank : 40) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
GATA6_MOUSE
|
||||||
| NC score | 0.055007 (rank : 41) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61169, P97729, Q9QZK3 | Gene names | Gata6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GATA6_HUMAN
|
||||||
| NC score | 0.054913 (rank : 42) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92908, P78327 | Gene names | GATA6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GATA5_HUMAN
|
||||||
| NC score | 0.051321 (rank : 43) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BWX5 | Gene names | GATA5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
K0146_MOUSE
|
||||||
| NC score | 0.040731 (rank : 44) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGX7, Q3TCD0, Q5U457, Q6A0B6, Q6IS60, Q811E1, Q8BWD6, Q8R305 | Gene names | Kiaa0146 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0146. | |||||
|
OFD1_HUMAN
|
||||||
| NC score | 0.040331 (rank : 45) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
RT09_HUMAN
|
||||||
| NC score | 0.033309 (rank : 46) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P82933 | Gene names | MRPS9, RPMS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 28S ribosomal protein S9, mitochondrial precursor (S9mt) (MRP-S9). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.030950 (rank : 47) | θ value | 0.0148317 (rank : 25) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
ARHGG_MOUSE
|
||||||
| NC score | 0.030415 (rank : 48) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3U5C8, Q501M8, Q8VCE8 | Gene names | Arhgef16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.030226 (rank : 49) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
UBP37_HUMAN
|
||||||
| NC score | 0.025413 (rank : 50) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.024683 (rank : 51) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.024433 (rank : 52) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
SWP70_HUMAN
|
||||||
| NC score | 0.024015 (rank : 53) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
CE110_HUMAN
|
||||||
| NC score | 0.022332 (rank : 54) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43303, O43335, Q68DV9, Q8NE13 | Gene names | CEP110, CP110, KIAA0419 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 110 kDa (Cep110 protein). | |||||
|
OM34_MOUSE
|
||||||
| NC score | 0.021208 (rank : 55) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CYG7, Q3THD0, Q9D059 | Gene names | Tomm34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.021077 (rank : 56) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
OM34_HUMAN
|
||||||
| NC score | 0.020356 (rank : 57) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15785, Q9NTZ3 | Gene names | TOMM34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit) (hTom34). | |||||
|
TPSNR_MOUSE
|
||||||
| NC score | 0.019232 (rank : 58) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VD31 | Gene names | Tapbpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tapasin-related protein precursor (TAPASIN-R) (Tapasin-like) (TAP- binding protein-related protein) (TAPBP-R) (TAP-binding protein-like). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.019209 (rank : 59) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.019064 (rank : 60) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
SKIL_MOUSE
|
||||||
| NC score | 0.018530 (rank : 61) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60665, Q60702, Q78E90, Q80VK5 | Gene names | Skil, Skir, Sno | |||
|
Domain Architecture |

|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.018502 (rank : 62) | θ value | 0.21417 (rank : 27) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
SPTN5_HUMAN
|
||||||
| NC score | 0.018231 (rank : 63) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
ARHGG_HUMAN
|
||||||
| NC score | 0.017588 (rank : 64) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VV41, Q86TF0, Q99434 | Gene names | ARHGEF16, NBR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.017564 (rank : 65) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
MYO9B_MOUSE
|
||||||
| NC score | 0.017430 (rank : 66) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
CTGE5_MOUSE
|
||||||
| NC score | 0.017184 (rank : 67) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 928 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R311, Q8CIE3 | Gene names | Ctage5, Mea6, Mgea6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 homolog (cTAGE-5 protein) (Meningioma-expressed antigen 6). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.016388 (rank : 68) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.015966 (rank : 69) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.015947 (rank : 70) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.015782 (rank : 71) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.015000 (rank : 72) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | 0.012104 (rank : 73) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
CHM4A_HUMAN
|
||||||
| NC score | 0.010650 (rank : 74) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BY43, Q32Q79, Q86SZ8, Q96QJ9, Q9P026 | Gene names | CHMP4A, C14orf123, SHAX2 | |||
|
Domain Architecture |

|
|||||
| Description | Charged multivesicular body protein 4a (Chromatin-modifying protein 4a) (CHMP4a) (Vacuolar protein sorting 7-1) (SNF7-1) (hSnf-1) (SNF7 homolog associated with Alix-2). | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.009778 (rank : 75) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
PPP5_HUMAN
|
||||||
| NC score | 0.009424 (rank : 76) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P53041, Q16722 | Gene names | PPP5C, PPP5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 5 (EC 3.1.3.16) (PP5) (Protein phosphatase T) (PP-T) (PPT). | |||||
|
ULK3_MOUSE
|
||||||
| NC score | 0.009195 (rank : 77) | θ value | 0.0431538 (rank : 26) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3U3Q1, Q8K1X6, Q9DBR8 | Gene names | Ulk3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK3 (EC 2.7.11.1) (Unc-51-like kinase 3). | |||||
|
STX2_HUMAN
|
||||||
| NC score | 0.008665 (rank : 78) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P32856, Q86VW8 | Gene names | STX2, EPIM | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-2 (Epimorphin). | |||||
|
AR13B_HUMAN
|
||||||
| NC score | 0.005551 (rank : 79) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3SXY8, Q504W8 | Gene names | ARL13B, ARL2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 13B (ADP-ribosylation factor-like protein 2-like 1) (ARL2-like protein 1). | |||||