Please be patient as the page loads
|
CAN3_MOUSE
|
||||||
| SwissProt Accessions | Q64691, Q9WUC5 | Gene names | Capn3 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CAN11_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.986356 (rank : 4) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UMQ6, Q8N4R5 | Gene names | CAPN11 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-11 (EC 3.4.22.-) (Calcium-activated neutral proteinase 11) (CANP 11). | |||||
|
CAN1_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.985679 (rank : 6) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P07384 | Gene names | CAPN1, CANPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
CAN1_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.985990 (rank : 5) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O35350, O88666 | Gene names | Capn1, Canp1, Capa1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
CAN2_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.985207 (rank : 8) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P17655, Q16738, Q6PJT3, Q8WU26, Q9HBB1 | Gene names | CAPN2, CANPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (Calpain large polypeptide L2). | |||||
|
CAN2_MOUSE
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.985530 (rank : 7) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08529, O35518, O54843 | Gene names | Capn2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (80 kDa M-calpain subunit) (CALP80). | |||||
|
CAN3_HUMAN
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.996198 (rank : 2) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P20807, Q9BTU4, Q9Y5S6, Q9Y5S7 | Gene names | CAPN3, CANP3, CANPL3, NCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3) (nCL-1). | |||||
|
CAN3_MOUSE
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q64691, Q9WUC5 | Gene names | Capn3 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3). | |||||
|
CAN9_HUMAN
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.987568 (rank : 3) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O14815, Q9NS74 | Gene names | CAPN9, NCL4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4) (CG36 protein). | |||||
|
CAN9_MOUSE
|
||||||
| θ value | 0 (rank : 9) | NC score | 0.985104 (rank : 9) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9D805, O35919 | Gene names | Capn9, Ncl4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4). | |||||
|
CAN12_HUMAN
|
||||||
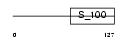
| θ value | 1.42914e-130 (rank : 10) | NC score | 0.968246 (rank : 10) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6ZSI9 | Gene names | CAPN12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain-12 (EC 3.4.22.-). | |||||
|
CAN12_MOUSE
|
||||||
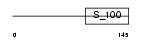
| θ value | 1.86652e-130 (rank : 11) | NC score | 0.967988 (rank : 11) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ER56, Q9ER53, Q9ER54, Q9ER55 | Gene names | Capn12 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-12 (EC 3.4.22.-). | |||||
|
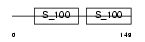
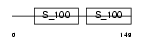
CAN5_MOUSE
|
||||||
| θ value | 1.54114e-100 (rank : 12) | NC score | 0.876250 (rank : 12) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08688, Q91YU0 | Gene names | Capn5 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-5 (EC 3.4.22.-) (nCL-3). | |||||
|
CAN5_HUMAN
|
||||||
| θ value | 4.48402e-100 (rank : 13) | NC score | 0.875701 (rank : 13) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15484, O00263 | Gene names | CAPN5 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-5 (EC 3.4.22.-) (nCL-3) (htra-3). | |||||
|
CAN6_HUMAN
|
||||||
| θ value | 1.03855e-80 (rank : 14) | NC score | 0.863824 (rank : 14) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6Q1, Q9UEQ1, Q9UJA8 | Gene names | CAPN6, CALPM, CANPX | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6 (Calpamodulin) (CalpM) (Calpain-like protease X-linked). | |||||
|
CAN6_MOUSE
|
||||||
| θ value | 1.26953e-78 (rank : 15) | NC score | 0.862866 (rank : 15) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35646 | Gene names | Capn6, Capa6 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6. | |||||
|
CPNS1_MOUSE
|
||||||
| θ value | 1.7238e-59 (rank : 16) | NC score | 0.834742 (rank : 18) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88456, Q3V0X5, Q5BKQ2, Q8CEI2, Q8VEK4, Q9R1C5 | Gene names | Capns1, Capn4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 1 (CSS1) (Calcium-dependent protease small subunit 1) (Calcium-dependent protease small subunit) (CDPS) (Calpain regulatory subunit) (Calcium-activated neutral proteinase small subunit) (CANP small subunit). | |||||
|
CPNS1_HUMAN
|
||||||
| θ value | 2.94036e-59 (rank : 17) | NC score | 0.833299 (rank : 19) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P04632, Q8WTX3, Q96EW0 | Gene names | CAPNS1, CAPN4, CAPNS | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 1 (CSS1) (Calcium-dependent protease small subunit 1) (Calcium-dependent protease small subunit) (CDPS) (Calpain regulatory subunit) (Calcium-activated neutral proteinase small subunit) (CANP small subunit). | |||||
|
CAN10_MOUSE
|
||||||
| θ value | 1.36585e-56 (rank : 18) | NC score | 0.848119 (rank : 16) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ESK3, Q99J13, Q9WVF0 | Gene names | Capn10, Capn8 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
CPNS2_MOUSE
|
||||||
| θ value | 2.57589e-55 (rank : 19) | NC score | 0.825844 (rank : 21) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9D7J7 | Gene names | Capns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
CPNS2_HUMAN
|
||||||
| θ value | 3.36421e-55 (rank : 20) | NC score | 0.829905 (rank : 20) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96L46, Q9BPV4 | Gene names | CAPNS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
CAN10_HUMAN
|
||||||
| θ value | 7.01481e-53 (rank : 21) | NC score | 0.844000 (rank : 17) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HC96, Q8NCD4, Q96IG4, Q96JI2, Q9HC89, Q9HC90, Q9HC91, Q9HC92, Q9HC93, Q9HC94, Q9HC95 | Gene names | CAPN10, KIAA1845 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
SORCN_HUMAN
|
||||||
| θ value | 1.86321e-21 (rank : 22) | NC score | 0.710882 (rank : 24) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P30626 | Gene names | SRI | |||
|
Domain Architecture |
|
|||||
| Description | Sorcin (22 kDa protein) (CP-22) (V19). | |||||
|
SORCN_MOUSE
|
||||||
| θ value | 1.86321e-21 (rank : 23) | NC score | 0.715455 (rank : 22) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6P069, Q3UKC5, Q9CR38, Q9D7V8 | Gene names | Sri | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorcin. | |||||
|
GRAN_MOUSE
|
||||||
| θ value | 9.24701e-21 (rank : 24) | NC score | 0.714887 (rank : 23) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8VC88, Q8BL53, Q8K3Z6 | Gene names | Gca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grancalcin. | |||||
|
GRAN_HUMAN
|
||||||
| θ value | 1.02238e-19 (rank : 25) | NC score | 0.707966 (rank : 25) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P28676 | Gene names | GCA, GCL | |||
|
Domain Architecture |
|
|||||
| Description | Grancalcin. | |||||
|
PDCD6_HUMAN
|
||||||
| θ value | 1.99529e-15 (rank : 26) | NC score | 0.598934 (rank : 30) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75340 | Gene names | PDCD6, ALG2 | |||
|
Domain Architecture |
|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2). | |||||
|
PDCD6_MOUSE
|
||||||
| θ value | 2.60593e-15 (rank : 27) | NC score | 0.596957 (rank : 31) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P12815, Q545I0, Q61145 | Gene names | Pdcd6, Alg2 | |||
|
Domain Architecture |
|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2) (PMP41) (ALG-257). | |||||
|
CAN7_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 28) | NC score | 0.644492 (rank : 29) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6W3 | Gene names | CAPN7, PALBH | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
CAN7_MOUSE
|
||||||
| θ value | 9.90251e-15 (rank : 29) | NC score | 0.646703 (rank : 28) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R1S8, Q9Z0P9 | Gene names | Capn7, Palbh | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
PEF1_HUMAN
|
||||||
| θ value | 1.29331e-14 (rank : 30) | NC score | 0.654447 (rank : 27) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UBV8 | Gene names | PEF1, ABP32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
PEF1_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 31) | NC score | 0.667490 (rank : 26) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BFY6, Q8VCT5, Q9CYW8, Q9D934 | Gene names | Pef1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
CALL5_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 32) | NC score | 0.207510 (rank : 32) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
CALM_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 33) | NC score | 0.135367 (rank : 34) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P62158, P02593, P70667, P99014, Q13942, Q53S29, Q61379, Q61380, Q96HK3 | Gene names | CALM1, CALM, CAM, CAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALM_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 34) | NC score | 0.135367 (rank : 35) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P62204, P02593, P70667, P99014, Q61379, Q61380 | Gene names | Calm1, Calm, Cam, Cam1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALM4_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 35) | NC score | 0.167854 (rank : 33) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
CALB1_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 36) | NC score | 0.110947 (rank : 47) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P05937 | Gene names | CALB1, CAB27 | |||
|
Domain Architecture |
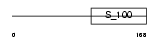
|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K). | |||||
|
CALB1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 37) | NC score | 0.115586 (rank : 42) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P12658, Q545N6 | Gene names | Calb1 | |||
|
Domain Architecture |
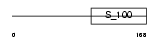
|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K) (Spot 35 protein) (PCD-29). | |||||
|
HM20B_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 38) | NC score | 0.034143 (rank : 77) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z104, Q9NSF7, Q9NSF8 | Gene names | Hmg20b, Braf35, Hmgx2, Smarce1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35). | |||||
|
CALL3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 39) | NC score | 0.128356 (rank : 38) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
SEGN_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 40) | NC score | 0.104287 (rank : 50) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O76038, Q5VV44, Q96QV7, Q9UJF6 | Gene names | SCGN, SECRET | |||
|
Domain Architecture |
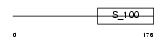
|
|||||
| Description | Secretagogin. | |||||
|
SEGN_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 41) | NC score | 0.092247 (rank : 52) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q91WD9 | Gene names | Scgn | |||
|
Domain Architecture |

|
|||||
| Description | Secretagogin. | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 42) | NC score | 0.012318 (rank : 99) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
VISL1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 43) | NC score | 0.029272 (rank : 80) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P62760, P28677, P29103, P42323, Q9UM20 | Gene names | VSNL1, VISL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Hippocalcin-like protein 3) (HLP3). | |||||
|
VISL1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 44) | NC score | 0.029272 (rank : 81) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P62761, P28677, P29103, P42323, Q9UM20 | Gene names | Vsnl1, Visl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Neural visinin-like protein 1) (NVL-1) (NVP-1). | |||||
|
HMGB1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 45) | NC score | 0.021661 (rank : 89) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09429, Q6IBE1 | Gene names | HMGB1, HMG1 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HMGB1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 46) | NC score | 0.021698 (rank : 88) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P63158, P07155, P27109, P27428 | Gene names | Hmgb1, Hmg-1, Hmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
CALL6_HUMAN
|
||||||
| θ value | 0.279714 (rank : 47) | NC score | 0.079947 (rank : 66) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TD86, Q6Q2C4 | Gene names | CALML6, CAGLP, CALGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-like protein 6 (Calglandulin-like protein). | |||||
|
EFHB_MOUSE
|
||||||
| θ value | 0.279714 (rank : 48) | NC score | 0.051510 (rank : 76) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CDU5 | Gene names | Efhb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
SPTA2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 49) | NC score | 0.029132 (rank : 82) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
TNNC2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 50) | NC score | 0.112801 (rank : 46) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P20801 | Gene names | Tnnc2, Tncs | |||
|
Domain Architecture |
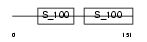
|
|||||
| Description | Troponin C, skeletal muscle (STNC). | |||||
|
CABP1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 51) | NC score | 0.091999 (rank : 53) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NZU7, O95663, Q9NZU8 | Gene names | CABP1 | |||
|
Domain Architecture |
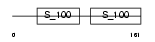
|
|||||
| Description | Calcium-binding protein 1 (CaBP1) (Calbrain). | |||||
|
CABP1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 52) | NC score | 0.092583 (rank : 51) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JLK7, Q9JLK6 | Gene names | Cabp1 | |||
|
Domain Architecture |
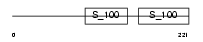
|
|||||
| Description | Calcium-binding protein 1 (CaBP1). | |||||
|
CANB2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 53) | NC score | 0.077235 (rank : 67) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q63811 | Gene names | Ppp3r2 | |||
|
Domain Architecture |
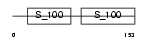
|
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform). | |||||
|
CTNL1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 54) | NC score | 0.023623 (rank : 86) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBT7, O76084, Q53FQ2, Q5JTQ7, Q5JTQ8, Q9Y401 | Gene names | CTNNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-catulin (Catenin alpha-like protein 1) (Alpha-catenin-related protein) (ACRP). | |||||
|
HM20B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 55) | NC score | 0.025885 (rank : 85) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P0W2, Q6IBP8, Q8NBD5, Q9HD21, Q9Y491, Q9Y4A2 | Gene names | HMG20B, BRAF35, HMGX2, SMARCE1R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35) (Sox-like transcriptional factor). | |||||
|
HNRPU_HUMAN
|
||||||
| θ value | 0.47712 (rank : 56) | NC score | 0.017972 (rank : 93) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q00839, O75507, Q96HY9 | Gene names | HNRPU, SAFA, U21.1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U (hnRNP U) (Scaffold attachment factor A) (SAF-A) (p120) (pp120). | |||||
|
TNNC2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 57) | NC score | 0.109015 (rank : 49) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P02585 | Gene names | TNNC2 | |||
|
Domain Architecture |
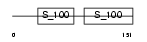
|
|||||
| Description | Troponin C, skeletal muscle. | |||||
|
CF103_HUMAN
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.131057 (rank : 37) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N7X0, Q5T402 | Gene names | C6orf103 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf103. | |||||
|
ASPH_HUMAN
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.022776 (rank : 87) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12797 | Gene names | ASPH | |||
|
Domain Architecture |

|
|||||
| Description | Aspartyl/asparaginyl beta-hydroxylase (EC 1.14.11.16) (Aspartate beta- hydroxylase) (ASP beta-hydroxylase) (Peptide-aspartate beta- dioxygenase). | |||||
|
EFHB_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.060426 (rank : 72) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8N7U6, Q6ZWK9, Q8IV58, Q96LQ6 | Gene names | EFHB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
CCAR1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.014517 (rank : 95) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | -0.001020 (rank : 132) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
DSG3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.003557 (rank : 120) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35902, Q8CB02, Q8CE48 | Gene names | Dsg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-3 precursor (130 kDa pemphigus vulgaris antigen homolog). | |||||
|
PLSI_HUMAN
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.088969 (rank : 56) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14651 | Gene names | PLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-1 (I-plastin) (Intestine-specific plastin). | |||||
|
BMP2K_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | -0.001620 (rank : 133) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91Z96, Q8C8L7 | Gene names | Bmp2k, Bike | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
CETN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.133621 (rank : 36) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q12798 | Gene names | CETN1, CEN1, CETN | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-1 (Caltractin isoform 2). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.007159 (rank : 108) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
NFL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.000805 (rank : 130) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 666 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P07196, Q16154, Q8IU72 | Gene names | NEFL, NF68, NFL | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.007358 (rank : 107) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
ACTN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.027918 (rank : 84) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P12814, Q9BTN1 | Gene names | ACTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
ACTN1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.028840 (rank : 83) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TPR4 | Gene names | Actn1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
CABP5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.082886 (rank : 65) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CCAR1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.014462 (rank : 96) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
CETN1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.121993 (rank : 40) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41209, Q3V119, Q9D9G9, Q9DAL6 | Gene names | Cetn1, Calt | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-1 (Caltractin). | |||||
|
FOXD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.001976 (rank : 126) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16676, Q12949 | Gene names | FOXD1, FKHL8, FREAC4 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein D1 (Forkhead-related protein FKHL8) (Forkhead- related transcription factor 4) (FREAC-4). | |||||
|
IF5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.012929 (rank : 97) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59325 | Gene names | Eif5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5 (eIF-5). | |||||
|
MYO6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.003143 (rank : 121) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64331 | Gene names | Myo6, Sv | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
PPE1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.032762 (rank : 79) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14829, O15253, Q9NU21, Q9UJH0 | Gene names | PPEF1, PPEF, PPP7C | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase with EF-hands 1 (EC 3.1.3.16) (PPEF-1) (Protein phosphatase with EF calcium-binding domain) (PPEF) (Serine/threonine-protein phosphatase 7) (PP7). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.004914 (rank : 113) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.004414 (rank : 117) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
E41L1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.004853 (rank : 115) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2H5 | Gene names | Epb41l1, Epb4, Epb4.1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
EFCB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.034077 (rank : 78) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HAE3 | Gene names | EFCAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
RYR2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.021460 (rank : 90) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
ACTN4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.020207 (rank : 92) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P57780 | Gene names | Actn4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-4 (Non-muscle alpha-actinin 4) (F-actin cross-linking protein). | |||||
|
E41L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.004466 (rank : 116) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
MYO6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.002151 (rank : 125) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
RPA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.010990 (rank : 101) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95602, Q9UEH0, Q9UFT9 | Gene names | POLR1A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
RPA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.010992 (rank : 100) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35134, Q7TSA9 | Gene names | Polr1a, Rpa1, Rpo1-4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
TNNC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.127272 (rank : 39) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P19123 | Gene names | Tnnc1, Tncc | |||
|
Domain Architecture |
|
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
TSH1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.008483 (rank : 104) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.008293 (rank : 105) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.008840 (rank : 102) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
TNNC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.121103 (rank : 41) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P63316, O14800, P02590, P04463 | Gene names | TNNC1, TNNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | -0.004382 (rank : 135) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
AF9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.005802 (rank : 111) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.002533 (rank : 123) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
CD2L1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | -0.004844 (rank : 136) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
DSG3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.001661 (rank : 127) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32926 | Gene names | DSG3 | |||
|
Domain Architecture |
|
|||||
| Description | Desmoglein-3 precursor (130 kDa pemphigus vulgaris antigen) (PVA). | |||||
|
ELF2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.007115 (rank : 109) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
MTMRD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.002918 (rank : 122) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86WG5, Q3MJF0, Q68DQ3, Q6P459, Q6PJD1, Q7Z325, Q7Z621, Q86VE2, Q96FE2, Q9C097 | Gene names | SBF2, CMT4B2, KIAA1766, MTMR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 13 (SET-binding factor 2). | |||||
|
NASP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.007832 (rank : 106) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
NMDE3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.001624 (rank : 128) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.000259 (rank : 131) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PRGC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.005129 (rank : 112) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
PTMA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.016419 (rank : 94) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
STAR8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.002439 (rank : 124) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
TECT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.006186 (rank : 110) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BZ64, Q3UMP0, Q8BUE2 | Gene names | Tect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tectonic-1 precursor. | |||||
|
ACTN4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.020437 (rank : 91) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43707, O76048 | Gene names | ACTN4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-4 (Non-muscle alpha-actinin 4) (F-actin cross-linking protein). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.001016 (rank : 129) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CN037_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.004903 (rank : 114) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.003855 (rank : 118) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
IF3I_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.008512 (rank : 103) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y262 | Gene names | EIF3S6IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 6-interacting protein. | |||||
|
KI21A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | -0.001902 (rank : 134) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.012757 (rank : 98) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
ZN318_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.003593 (rank : 119) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
CABP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.083476 (rank : 64) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NPB3 | Gene names | CABP2 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CABP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.086588 (rank : 61) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JLK4, Q9JLK5 | Gene names | Cabp2 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CABP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.063560 (rank : 71) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZU6, Q9NYY0, Q9NZT9 | Gene names | CABP3 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 3 (CaBP3). | |||||
|
CABP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.114688 (rank : 44) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CABP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.085020 (rank : 63) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CABP5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.067223 (rank : 70) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CABP7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.088503 (rank : 57) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86V35 | Gene names | CABP7 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CABP7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.088503 (rank : 58) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91ZM8 | Gene names | Cabp7 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CALB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.112944 (rank : 45) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
CALB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.085787 (rank : 62) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
CALN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.056359 (rank : 73) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BXU9 | Gene names | CALN1, CABP8 | |||
|
Domain Architecture |
|
|||||
| Description | Calneuron-1 (Calcium-binding protein CaBP8). | |||||
|
CALN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.056230 (rank : 74) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JJG7 | Gene names | Caln1 | |||
|
Domain Architecture |
|
|||||
| Description | Calneuron-1. | |||||
|
CANB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.087195 (rank : 59) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P63098, P06705, P15117, Q08044 | Gene names | PPP3R1, CNA2, CNB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.087192 (rank : 60) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q63810 | Gene names | Ppp3r1, Cnb | |||
|
Domain Architecture |
|
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.074155 (rank : 68) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96LZ3, Q7Z4V8, Q8WYJ4 | Gene names | PPP3R2, CBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform) (Calcineurin B-like protein) (CBLP) (CNBII). | |||||
|
CAYP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.072402 (rank : 69) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13938, Q3MJA7, Q8WUZ3 | Gene names | CAPS | |||
|
Domain Architecture |
|
|||||
| Description | Calcyphosin (Calcyphosine). | |||||
|
CETN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.110284 (rank : 48) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41208, Q53XW1 | Gene names | CETN2, CALT, CEN2 | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CETN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.114938 (rank : 43) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CETN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.091443 (rank : 55) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O15182, Q9BS23 | Gene names | CETN3, CEN3 | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-3. | |||||
|
CETN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.091683 (rank : 54) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O35648 | Gene names | Cetn3, Cen3 | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-3. | |||||
|
PRVA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.055180 (rank : 75) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P32848 | Gene names | Pvalb, Pva | |||
|
Domain Architecture |
|
|||||
| Description | Parvalbumin alpha. | |||||
|
CAN3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q64691, Q9WUC5 | Gene names | Capn3 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3). | |||||
|
CAN3_HUMAN
|
||||||
| NC score | 0.996198 (rank : 2) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P20807, Q9BTU4, Q9Y5S6, Q9Y5S7 | Gene names | CAPN3, CANP3, CANPL3, NCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3) (nCL-1). | |||||
|
CAN9_HUMAN
|
||||||
| NC score | 0.987568 (rank : 3) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O14815, Q9NS74 | Gene names | CAPN9, NCL4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4) (CG36 protein). | |||||
|
CAN11_HUMAN
|
||||||
| NC score | 0.986356 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UMQ6, Q8N4R5 | Gene names | CAPN11 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-11 (EC 3.4.22.-) (Calcium-activated neutral proteinase 11) (CANP 11). | |||||
|
CAN1_MOUSE
|
||||||
| NC score | 0.985990 (rank : 5) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O35350, O88666 | Gene names | Capn1, Canp1, Capa1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
CAN1_HUMAN
|
||||||
| NC score | 0.985679 (rank : 6) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P07384 | Gene names | CAPN1, CANPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
CAN2_MOUSE
|
||||||
| NC score | 0.985530 (rank : 7) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08529, O35518, O54843 | Gene names | Capn2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (80 kDa M-calpain subunit) (CALP80). | |||||
|
CAN2_HUMAN
|
||||||
| NC score | 0.985207 (rank : 8) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P17655, Q16738, Q6PJT3, Q8WU26, Q9HBB1 | Gene names | CAPN2, CANPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (Calpain large polypeptide L2). | |||||
|
CAN9_MOUSE
|
||||||
| NC score | 0.985104 (rank : 9) | θ value | 0 (rank : 9) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9D805, O35919 | Gene names | Capn9, Ncl4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4). | |||||
|
CAN12_HUMAN
|
||||||
| NC score | 0.968246 (rank : 10) | θ value | 1.42914e-130 (rank : 10) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6ZSI9 | Gene names | CAPN12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain-12 (EC 3.4.22.-). | |||||
|
CAN12_MOUSE
|
||||||
| NC score | 0.967988 (rank : 11) | θ value | 1.86652e-130 (rank : 11) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ER56, Q9ER53, Q9ER54, Q9ER55 | Gene names | Capn12 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-12 (EC 3.4.22.-). | |||||
|
CAN5_MOUSE
|
||||||
| NC score | 0.876250 (rank : 12) | θ value | 1.54114e-100 (rank : 12) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08688, Q91YU0 | Gene names | Capn5 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-5 (EC 3.4.22.-) (nCL-3). | |||||
|
CAN5_HUMAN
|
||||||
| NC score | 0.875701 (rank : 13) | θ value | 4.48402e-100 (rank : 13) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15484, O00263 | Gene names | CAPN5 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-5 (EC 3.4.22.-) (nCL-3) (htra-3). | |||||
|
CAN6_HUMAN
|
||||||
| NC score | 0.863824 (rank : 14) | θ value | 1.03855e-80 (rank : 14) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6Q1, Q9UEQ1, Q9UJA8 | Gene names | CAPN6, CALPM, CANPX | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6 (Calpamodulin) (CalpM) (Calpain-like protease X-linked). | |||||
|
CAN6_MOUSE
|
||||||
| NC score | 0.862866 (rank : 15) | θ value | 1.26953e-78 (rank : 15) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35646 | Gene names | Capn6, Capa6 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6. | |||||
|
CAN10_MOUSE
|
||||||
| NC score | 0.848119 (rank : 16) | θ value | 1.36585e-56 (rank : 18) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ESK3, Q99J13, Q9WVF0 | Gene names | Capn10, Capn8 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
CAN10_HUMAN
|
||||||
| NC score | 0.844000 (rank : 17) | θ value | 7.01481e-53 (rank : 21) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HC96, Q8NCD4, Q96IG4, Q96JI2, Q9HC89, Q9HC90, Q9HC91, Q9HC92, Q9HC93, Q9HC94, Q9HC95 | Gene names | CAPN10, KIAA1845 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
CPNS1_MOUSE
|
||||||
| NC score | 0.834742 (rank : 18) | θ value | 1.7238e-59 (rank : 16) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88456, Q3V0X5, Q5BKQ2, Q8CEI2, Q8VEK4, Q9R1C5 | Gene names | Capns1, Capn4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 1 (CSS1) (Calcium-dependent protease small subunit 1) (Calcium-dependent protease small subunit) (CDPS) (Calpain regulatory subunit) (Calcium-activated neutral proteinase small subunit) (CANP small subunit). | |||||
|
CPNS1_HUMAN
|
||||||
| NC score | 0.833299 (rank : 19) | θ value | 2.94036e-59 (rank : 17) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P04632, Q8WTX3, Q96EW0 | Gene names | CAPNS1, CAPN4, CAPNS | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 1 (CSS1) (Calcium-dependent protease small subunit 1) (Calcium-dependent protease small subunit) (CDPS) (Calpain regulatory subunit) (Calcium-activated neutral proteinase small subunit) (CANP small subunit). | |||||
|
CPNS2_HUMAN
|
||||||
| NC score | 0.829905 (rank : 20) | θ value | 3.36421e-55 (rank : 20) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96L46, Q9BPV4 | Gene names | CAPNS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
CPNS2_MOUSE
|
||||||
| NC score | 0.825844 (rank : 21) | θ value | 2.57589e-55 (rank : 19) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9D7J7 | Gene names | Capns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
SORCN_MOUSE
|
||||||
| NC score | 0.715455 (rank : 22) | θ value | 1.86321e-21 (rank : 23) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6P069, Q3UKC5, Q9CR38, Q9D7V8 | Gene names | Sri | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorcin. | |||||
|
GRAN_MOUSE
|
||||||
| NC score | 0.714887 (rank : 23) | θ value | 9.24701e-21 (rank : 24) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8VC88, Q8BL53, Q8K3Z6 | Gene names | Gca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grancalcin. | |||||
|
SORCN_HUMAN
|
||||||
| NC score | 0.710882 (rank : 24) | θ value | 1.86321e-21 (rank : 22) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P30626 | Gene names | SRI | |||
|
Domain Architecture |

|
|||||
| Description | Sorcin (22 kDa protein) (CP-22) (V19). | |||||
|
GRAN_HUMAN
|
||||||
| NC score | 0.707966 (rank : 25) | θ value | 1.02238e-19 (rank : 25) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P28676 | Gene names | GCA, GCL | |||
|
Domain Architecture |

|
|||||
| Description | Grancalcin. | |||||
|
PEF1_MOUSE
|
||||||
| NC score | 0.667490 (rank : 26) | θ value | 2.20605e-14 (rank : 31) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BFY6, Q8VCT5, Q9CYW8, Q9D934 | Gene names | Pef1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
PEF1_HUMAN
|
||||||
| NC score | 0.654447 (rank : 27) | θ value | 1.29331e-14 (rank : 30) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UBV8 | Gene names | PEF1, ABP32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
CAN7_MOUSE
|
||||||
| NC score | 0.646703 (rank : 28) | θ value | 9.90251e-15 (rank : 29) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R1S8, Q9Z0P9 | Gene names | Capn7, Palbh | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
CAN7_HUMAN
|
||||||
| NC score | 0.644492 (rank : 29) | θ value | 9.90251e-15 (rank : 28) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6W3 | Gene names | CAPN7, PALBH | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
PDCD6_HUMAN
|
||||||
| NC score | 0.598934 (rank : 30) | θ value | 1.99529e-15 (rank : 26) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75340 | Gene names | PDCD6, ALG2 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2). | |||||
|
PDCD6_MOUSE
|
||||||
| NC score | 0.596957 (rank : 31) | θ value | 2.60593e-15 (rank : 27) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P12815, Q545I0, Q61145 | Gene names | Pdcd6, Alg2 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2) (PMP41) (ALG-257). | |||||
|
CALL5_HUMAN
|
||||||
| NC score | 0.207510 (rank : 32) | θ value | 1.43324e-05 (rank : 32) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
CALM4_MOUSE
|
||||||
| NC score | 0.167854 (rank : 33) | θ value | 0.00134147 (rank : 35) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
CALM_HUMAN
|
||||||
| NC score | 0.135367 (rank : 34) | θ value | 0.000461057 (rank : 33) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P62158, P02593, P70667, P99014, Q13942, Q53S29, Q61379, Q61380, Q96HK3 | Gene names | CALM1, CALM, CAM, CAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALM_MOUSE
|
||||||
| NC score | 0.135367 (rank : 35) | θ value | 0.000461057 (rank : 34) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P62204, P02593, P70667, P99014, Q61379, Q61380 | Gene names | Calm1, Calm, Cam, Cam1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CETN1_HUMAN
|
||||||
| NC score | 0.133621 (rank : 36) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q12798 | Gene names | CETN1, CEN1, CETN | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-1 (Caltractin isoform 2). | |||||
|
CF103_HUMAN
|
||||||
| NC score | 0.131057 (rank : 37) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N7X0, Q5T402 | Gene names | C6orf103 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf103. | |||||
|
CALL3_HUMAN
|
||||||
| NC score | 0.128356 (rank : 38) | θ value | 0.0563607 (rank : 39) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
TNNC1_MOUSE
|
||||||
| NC score | 0.127272 (rank : 39) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P19123 | Gene names | Tnnc1, Tncc | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
CETN1_MOUSE
|
||||||
| NC score | 0.121993 (rank : 40) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41209, Q3V119, Q9D9G9, Q9DAL6 | Gene names | Cetn1, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-1 (Caltractin). | |||||
|
TNNC1_HUMAN
|
||||||
| NC score | 0.121103 (rank : 41) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P63316, O14800, P02590, P04463 | Gene names | TNNC1, TNNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
CALB1_MOUSE
|
||||||
| NC score | 0.115586 (rank : 42) | θ value | 0.00509761 (rank : 37) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P12658, Q545N6 | Gene names | Calb1 | |||
|
Domain Architecture |

|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K) (Spot 35 protein) (PCD-29). | |||||
|
CETN2_MOUSE
|
||||||
| NC score | 0.114938 (rank : 43) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CABP4_HUMAN
|
||||||
| NC score | 0.114688 (rank : 44) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CALB2_HUMAN
|
||||||
| NC score | 0.112944 (rank : 45) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
TNNC2_MOUSE
|
||||||
| NC score | 0.112801 (rank : 46) | θ value | 0.279714 (rank : 50) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P20801 | Gene names | Tnnc2, Tncs | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, skeletal muscle (STNC). | |||||
|
CALB1_HUMAN
|
||||||
| NC score | 0.110947 (rank : 47) | θ value | 0.00509761 (rank : 36) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P05937 | Gene names | CALB1, CAB27 | |||
|
Domain Architecture |

|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K). | |||||
|
CETN2_HUMAN
|
||||||
| NC score | 0.110284 (rank : 48) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41208, Q53XW1 | Gene names | CETN2, CALT, CEN2 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
TNNC2_HUMAN
|
||||||
| NC score | 0.109015 (rank : 49) | θ value | 0.47712 (rank : 57) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P02585 | Gene names | TNNC2 | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, skeletal muscle. | |||||
|
SEGN_HUMAN
|
||||||
| NC score | 0.104287 (rank : 50) | θ value | 0.0961366 (rank : 40) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O76038, Q5VV44, Q96QV7, Q9UJF6 | Gene names | SCGN, SECRET | |||
|
Domain Architecture |

|
|||||
| Description | Secretagogin. | |||||
|
CABP1_MOUSE
|
||||||
| NC score | 0.092583 (rank : 51) | θ value | 0.365318 (rank : 52) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JLK7, Q9JLK6 | Gene names | Cabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 1 (CaBP1). | |||||
|
SEGN_MOUSE
|
||||||
| NC score | 0.092247 (rank : 52) | θ value | 0.0961366 (rank : 41) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q91WD9 | Gene names | Scgn | |||
|
Domain Architecture |

|
|||||
| Description | Secretagogin. | |||||
|
CABP1_HUMAN
|
||||||
| NC score | 0.091999 (rank : 53) | θ value | 0.365318 (rank : 51) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NZU7, O95663, Q9NZU8 | Gene names | CABP1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 1 (CaBP1) (Calbrain). | |||||
|
CETN3_MOUSE
|
||||||
| NC score | 0.091683 (rank : 54) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O35648 | Gene names | Cetn3, Cen3 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-3. | |||||
|
CETN3_HUMAN
|
||||||
| NC score | 0.091443 (rank : 55) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O15182, Q9BS23 | Gene names | CETN3, CEN3 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-3. | |||||
|
PLSI_HUMAN
|
||||||
| NC score | 0.088969 (rank : 56) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14651 | Gene names | PLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-1 (I-plastin) (Intestine-specific plastin). | |||||
|
CABP7_HUMAN
|
||||||
| NC score | 0.088503 (rank : 57) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86V35 | Gene names | CABP7 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CABP7_MOUSE
|
||||||
| NC score | 0.088503 (rank : 58) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91ZM8 | Gene names | Cabp7 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CANB1_HUMAN
|
||||||
| NC score | 0.087195 (rank : 59) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P63098, P06705, P15117, Q08044 | Gene names | PPP3R1, CNA2, CNB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB1_MOUSE
|
||||||
| NC score | 0.087192 (rank : 60) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q63810 | Gene names | Ppp3r1, Cnb | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CABP2_MOUSE
|
||||||
| NC score | 0.086588 (rank : 61) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JLK4, Q9JLK5 | Gene names | Cabp2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CALB2_MOUSE
|
||||||
| NC score | 0.085787 (rank : 62) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
CABP4_MOUSE
|
||||||
| NC score | 0.085020 (rank : 63) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CABP2_HUMAN
|
||||||
| NC score | 0.083476 (rank : 64) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NPB3 | Gene names | CABP2 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CABP5_HUMAN
|
||||||
| NC score | 0.082886 (rank : 65) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CALL6_HUMAN
|
||||||
| NC score | 0.079947 (rank : 66) | θ value | 0.279714 (rank : 47) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TD86, Q6Q2C4 | Gene names | CALML6, CAGLP, CALGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-like protein 6 (Calglandulin-like protein). | |||||
|
CANB2_MOUSE
|
||||||
| NC score | 0.077235 (rank : 67) | θ value | 0.365318 (rank : 53) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q63811 | Gene names | Ppp3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform). | |||||
|
CANB2_HUMAN
|
||||||
| NC score | 0.074155 (rank : 68) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96LZ3, Q7Z4V8, Q8WYJ4 | Gene names | PPP3R2, CBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform) (Calcineurin B-like protein) (CBLP) (CNBII). | |||||
|
CAYP1_HUMAN
|
||||||
| NC score | 0.072402 (rank : 69) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13938, Q3MJA7, Q8WUZ3 | Gene names | CAPS | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin (Calcyphosine). | |||||
|
CABP5_MOUSE
|
||||||
| NC score | 0.067223 (rank : 70) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CABP3_HUMAN
|
||||||
| NC score | 0.063560 (rank : 71) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZU6, Q9NYY0, Q9NZT9 | Gene names | CABP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 3 (CaBP3). | |||||
|
EFHB_HUMAN
|
||||||
| NC score | 0.060426 (rank : 72) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8N7U6, Q6ZWK9, Q8IV58, Q96LQ6 | Gene names | EFHB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
CALN1_HUMAN
|
||||||
| NC score | 0.056359 (rank : 73) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BXU9 | Gene names | CALN1, CABP8 | |||
|
Domain Architecture |

|
|||||
| Description | Calneuron-1 (Calcium-binding protein CaBP8). | |||||
|
CALN1_MOUSE
|
||||||
| NC score | 0.056230 (rank : 74) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JJG7 | Gene names | Caln1 | |||
|
Domain Architecture |

|
|||||
| Description | Calneuron-1. | |||||
|
PRVA_MOUSE
|
||||||
| NC score | 0.055180 (rank : 75) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P32848 | Gene names | Pvalb, Pva | |||
|
Domain Architecture |

|
|||||
| Description | Parvalbumin alpha. | |||||
|
EFHB_MOUSE
|
||||||
| NC score | 0.051510 (rank : 76) | θ value | 0.279714 (rank : 48) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CDU5 | Gene names | Efhb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
HM20B_MOUSE
|
||||||
| NC score | 0.034143 (rank : 77) | θ value | 0.00509761 (rank : 38) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z104, Q9NSF7, Q9NSF8 | Gene names | Hmg20b, Braf35, Hmgx2, Smarce1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35). | |||||
|
EFCB1_HUMAN
|
||||||
| NC score | 0.034077 (rank : 78) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HAE3 | Gene names | EFCAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
PPE1_HUMAN
|
||||||
| NC score | 0.032762 (rank : 79) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14829, O15253, Q9NU21, Q9UJH0 | Gene names | PPEF1, PPEF, PPP7C | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase with EF-hands 1 (EC 3.1.3.16) (PPEF-1) (Protein phosphatase with EF calcium-binding domain) (PPEF) (Serine/threonine-protein phosphatase 7) (PP7). | |||||
|
VISL1_HUMAN
|
||||||
| NC score | 0.029272 (rank : 80) | θ value | 0.125558 (rank : 43) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P62760, P28677, P29103, P42323, Q9UM20 | Gene names | VSNL1, VISL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Hippocalcin-like protein 3) (HLP3). | |||||
|
VISL1_MOUSE
|
||||||
| NC score | 0.029272 (rank : 81) | θ value | 0.125558 (rank : 44) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P62761, P28677, P29103, P42323, Q9UM20 | Gene names | Vsnl1, Visl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Neural visinin-like protein 1) (NVL-1) (NVP-1). | |||||
|
SPTA2_HUMAN
|
||||||
| NC score | 0.029132 (rank : 82) | θ value | 0.279714 (rank : 49) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
ACTN1_MOUSE
|
||||||
| NC score | 0.028840 (rank : 83) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TPR4 | Gene names | Actn1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
ACTN1_HUMAN
|
||||||
| NC score | 0.027918 (rank : 84) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P12814, Q9BTN1 | Gene names | ACTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
HM20B_HUMAN
|
||||||
| NC score | 0.025885 (rank : 85) | θ value | 0.47712 (rank : 55) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P0W2, Q6IBP8, Q8NBD5, Q9HD21, Q9Y491, Q9Y4A2 | Gene names | HMG20B, BRAF35, HMGX2, SMARCE1R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35) (Sox-like transcriptional factor). | |||||
|
CTNL1_HUMAN
|
||||||
| NC score | 0.023623 (rank : 86) | θ value | 0.47712 (rank : 54) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBT7, O76084, Q53FQ2, Q5JTQ7, Q5JTQ8, Q9Y401 | Gene names | CTNNAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-catulin (Catenin alpha-like protein 1) (Alpha-catenin-related protein) (ACRP). | |||||
|
ASPH_HUMAN
|
||||||
| NC score | 0.022776 (rank : 87) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12797 | Gene names | ASPH | |||
|
Domain Architecture |

|
|||||
| Description | Aspartyl/asparaginyl beta-hydroxylase (EC 1.14.11.16) (Aspartate beta- hydroxylase) (ASP beta-hydroxylase) (Peptide-aspartate beta- dioxygenase). | |||||
|
HMGB1_MOUSE
|
||||||
| NC score | 0.021698 (rank : 88) | θ value | 0.163984 (rank : 46) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P63158, P07155, P27109, P27428 | Gene names | Hmgb1, Hmg-1, Hmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HMGB1_HUMAN
|
||||||
| NC score | 0.021661 (rank : 89) | θ value | 0.163984 (rank : 45) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09429, Q6IBE1 | Gene names | HMGB1, HMG1 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
RYR2_HUMAN
|
||||||
| NC score | 0.021460 (rank : 90) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
ACTN4_HUMAN
|
||||||
| NC score | 0.020437 (rank : 91) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43707, O76048 | Gene names | ACTN4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-4 (Non-muscle alpha-actinin 4) (F-actin cross-linking protein). | |||||
|
ACTN4_MOUSE
|
||||||
| NC score | 0.020207 (rank : 92) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P57780 | Gene names | Actn4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-4 (Non-muscle alpha-actinin 4) (F-actin cross-linking protein). | |||||
|
HNRPU_HUMAN
|
||||||
| NC score | 0.017972 (rank : 93) | θ value | 0.47712 (rank : 56) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q00839, O75507, Q96HY9 | Gene names | HNRPU, SAFA, U21.1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U (hnRNP U) (Scaffold attachment factor A) (SAF-A) (p120) (pp120). | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.016419 (rank : 94) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
CCAR1_HUMAN
|
||||||
| NC score | 0.014517 (rank : 95) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
CCAR1_MOUSE
|
||||||
| NC score | 0.014462 (rank : 96) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
IF5_MOUSE
|
||||||
| NC score | 0.012929 (rank : 97) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59325 | Gene names | Eif5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5 (eIF-5). | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.012757 (rank : 98) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.012318 (rank : 99) | θ value | 0.125558 (rank : 42) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
RPA1_MOUSE
|
||||||
| NC score | 0.010992 (rank : 100) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35134, Q7TSA9 | Gene names | Polr1a, Rpa1, Rpo1-4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
RPA1_HUMAN
|
||||||
| NC score | 0.010990 (rank : 101) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95602, Q9UEH0, Q9UFT9 | Gene names | POLR1A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.008840 (rank : 102) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
IF3I_HUMAN
|
||||||
| NC score | 0.008512 (rank : 103) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y262 | Gene names | EIF3S6IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 6-interacting protein. | |||||
|
TSH1_MOUSE
|
||||||
| NC score | 0.008483 (rank : 104) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.008293 (rank : 105) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.007832 (rank : 106) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.007358 (rank : 107) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.007159 (rank : 108) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
ELF2_MOUSE
|
||||||
| NC score | 0.007115 (rank : 109) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
TECT1_MOUSE
|
||||||
| NC score | 0.006186 (rank : 110) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BZ64, Q3UMP0, Q8BUE2 | Gene names | Tect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tectonic-1 precursor. | |||||
|
AF9_HUMAN
|
||||||
| NC score | 0.005802 (rank : 111) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
PRGC2_MOUSE
|
||||||
| NC score | 0.005129 (rank : 112) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.004914 (rank : 113) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
CN037_MOUSE
|
||||||
| NC score | 0.004903 (rank : 114) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
E41L1_MOUSE
|
||||||
| NC score | 0.004853 (rank : 115) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2H5 | Gene names | Epb41l1, Epb4, Epb4.1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
E41L1_HUMAN
|
||||||
| NC score | 0.004466 (rank : 116) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.004414 (rank : 117) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.003855 (rank : 118) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
ZN318_HUMAN
|
||||||
| NC score | 0.003593 (rank : 119) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
DSG3_MOUSE
|
||||||
| NC score | 0.003557 (rank : 120) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35902, Q8CB02, Q8CE48 | Gene names | Dsg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-3 precursor (130 kDa pemphigus vulgaris antigen homolog). | |||||
|
MYO6_MOUSE
|
||||||
| NC score | 0.003143 (rank : 121) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64331 | Gene names | Myo6, Sv | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
MTMRD_HUMAN
|
||||||
| NC score | 0.002918 (rank : 122) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86WG5, Q3MJF0, Q68DQ3, Q6P459, Q6PJD1, Q7Z325, Q7Z621, Q86VE2, Q96FE2, Q9C097 | Gene names | SBF2, CMT4B2, KIAA1766, MTMR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 13 (SET-binding factor 2). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.002533 (rank : 123) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
STAR8_HUMAN
|
||||||
| NC score | 0.002439 (rank : 124) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
MYO6_HUMAN
|
||||||
| NC score | 0.002151 (rank : 125) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
FOXD1_HUMAN
|
||||||
| NC score | 0.001976 (rank : 126) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16676, Q12949 | Gene names | FOXD1, FKHL8, FREAC4 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D1 (Forkhead-related protein FKHL8) (Forkhead- related transcription factor 4) (FREAC-4). | |||||
|
DSG3_HUMAN
|
||||||
| NC score | 0.001661 (rank : 127) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32926 | Gene names | DSG3 | |||
|
Domain Architecture |

|
|||||
| Description | Desmoglein-3 precursor (130 kDa pemphigus vulgaris antigen) (PVA). | |||||
|
NMDE3_HUMAN
|
||||||
| NC score | 0.001624 (rank : 128) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.001016 (rank : 129) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
NFL_HUMAN
|
||||||
| NC score | 0.000805 (rank : 130) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 666 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P07196, Q16154, Q8IU72 | Gene names | NEFL, NF68, NFL | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.000259 (rank : 131) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | -0.001020 (rank : 132) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
BMP2K_MOUSE
|
||||||
| NC score | -0.001620 (rank : 133) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91Z96, Q8C8L7 | Gene names | Bmp2k, Bike | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
KI21A_HUMAN
|
||||||
| NC score | -0.001902 (rank : 134) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | -0.004382 (rank : 135) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
CD2L1_MOUSE
|
||||||
| NC score | -0.004844 (rank : 136) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||