Please be patient as the page loads
|
CAN6_MOUSE
|
||||||
| SwissProt Accessions | O35646 | Gene names | Capn6, Capa6 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CAN6_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999578 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y6Q1, Q9UEQ1, Q9UJA8 | Gene names | CAPN6, CALPM, CANPX | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6 (Calpamodulin) (CalpM) (Calpain-like protease X-linked). | |||||
|
CAN6_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35646 | Gene names | Capn6, Capa6 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6. | |||||
|
CAN5_MOUSE
|
||||||
| θ value | 5.36813e-170 (rank : 3) | NC score | 0.987233 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08688, Q91YU0 | Gene names | Capn5 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-5 (EC 3.4.22.-) (nCL-3). | |||||
|
CAN5_HUMAN
|
||||||
| θ value | 3.47952e-169 (rank : 4) | NC score | 0.987572 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15484, O00263 | Gene names | CAPN5 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-5 (EC 3.4.22.-) (nCL-3) (htra-3). | |||||
|
CAN9_MOUSE
|
||||||
| θ value | 7.19216e-82 (rank : 5) | NC score | 0.884082 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D805, O35919 | Gene names | Capn9, Ncl4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4). | |||||
|
CAN9_HUMAN
|
||||||
| θ value | 5.69867e-79 (rank : 6) | NC score | 0.883968 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14815, Q9NS74 | Gene names | CAPN9, NCL4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4) (CG36 protein). | |||||
|
CAN3_MOUSE
|
||||||
| θ value | 1.26953e-78 (rank : 7) | NC score | 0.862866 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64691, Q9WUC5 | Gene names | Capn3 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3). | |||||
|
CAN2_HUMAN
|
||||||
| θ value | 2.1655e-78 (rank : 8) | NC score | 0.881548 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17655, Q16738, Q6PJT3, Q8WU26, Q9HBB1 | Gene names | CAPN2, CANPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (Calpain large polypeptide L2). | |||||
|
CAN2_MOUSE
|
||||||
| θ value | 2.82823e-78 (rank : 9) | NC score | 0.879035 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08529, O35518, O54843 | Gene names | Capn2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (80 kDa M-calpain subunit) (CALP80). | |||||
|
CAN3_HUMAN
|
||||||
| θ value | 3.12697e-77 (rank : 10) | NC score | 0.866187 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20807, Q9BTU4, Q9Y5S6, Q9Y5S7 | Gene names | CAPN3, CANP3, CANPL3, NCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3) (nCL-1). | |||||
|
CAN1_MOUSE
|
||||||
| θ value | 3.23591e-74 (rank : 11) | NC score | 0.871100 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35350, O88666 | Gene names | Capn1, Canp1, Capa1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
CAN11_HUMAN
|
||||||
| θ value | 3.34864e-71 (rank : 12) | NC score | 0.882701 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UMQ6, Q8N4R5 | Gene names | CAPN11 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-11 (EC 3.4.22.-) (Calcium-activated neutral proteinase 11) (CANP 11). | |||||
|
CAN1_HUMAN
|
||||||
| θ value | 1.66191e-70 (rank : 13) | NC score | 0.869982 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P07384 | Gene names | CAPN1, CANPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
CAN12_MOUSE
|
||||||
| θ value | 1.07972e-61 (rank : 14) | NC score | 0.901330 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ER56, Q9ER53, Q9ER54, Q9ER55 | Gene names | Capn12 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-12 (EC 3.4.22.-). | |||||
|
CAN12_HUMAN
|
||||||
| θ value | 8.55514e-59 (rank : 15) | NC score | 0.902450 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZSI9 | Gene names | CAPN12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain-12 (EC 3.4.22.-). | |||||
|
CAN10_MOUSE
|
||||||
| θ value | 3.48949e-44 (rank : 16) | NC score | 0.918285 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ESK3, Q99J13, Q9WVF0 | Gene names | Capn10, Capn8 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
CAN10_HUMAN
|
||||||
| θ value | 3.61106e-41 (rank : 17) | NC score | 0.914336 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HC96, Q8NCD4, Q96IG4, Q96JI2, Q9HC89, Q9HC90, Q9HC91, Q9HC92, Q9HC93, Q9HC94, Q9HC95 | Gene names | CAPN10, KIAA1845 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
CAN7_HUMAN
|
||||||
| θ value | 1.86321e-21 (rank : 18) | NC score | 0.750222 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6W3 | Gene names | CAPN7, PALBH | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
CAN7_MOUSE
|
||||||
| θ value | 2.43343e-21 (rank : 19) | NC score | 0.751242 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R1S8, Q9Z0P9 | Gene names | Capn7, Palbh | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
CF103_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 20) | NC score | 0.182148 (rank : 32) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N7X0, Q5T402 | Gene names | C6orf103 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf103. | |||||
|
RFIP2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.051922 (rank : 35) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7L804, Q9Y2F0 | Gene names | RAB11FIP2, KIAA0941 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 2 (Rab11-FIP2) (NRip11). | |||||
|
UN13A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.043333 (rank : 37) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
UN13B_MOUSE
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.034819 (rank : 38) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z1N9 | Gene names | Unc13b, Unc13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
RFIP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.027524 (rank : 40) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D620, Q3UBC2, Q8BN24 | Gene names | Rab11fip1, Rcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
UN13B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.029082 (rank : 39) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14795 | Gene names | UNC13B, UNC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
RFIP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.023191 (rank : 41) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
ZN292_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.003224 (rank : 43) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||
|
NIBA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.013999 (rank : 42) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZQ8, Q5TEM8, Q8TEI5, Q9H593, Q9H9Y8, Q9HCB9 | Gene names | C1orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban protein. | |||||
|
CALL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.080753 (rank : 33) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
CALM4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.061183 (rank : 34) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
CPNS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.479219 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P04632, Q8WTX3, Q96EW0 | Gene names | CAPNS1, CAPN4, CAPNS | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 1 (CSS1) (Calcium-dependent protease small subunit 1) (Calcium-dependent protease small subunit) (CDPS) (Calpain regulatory subunit) (Calcium-activated neutral proteinase small subunit) (CANP small subunit). | |||||
|
CPNS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.482685 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88456, Q3V0X5, Q5BKQ2, Q8CEI2, Q8VEK4, Q9R1C5 | Gene names | Capns1, Capn4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 1 (CSS1) (Calcium-dependent protease small subunit 1) (Calcium-dependent protease small subunit) (CDPS) (Calpain regulatory subunit) (Calcium-activated neutral proteinase small subunit) (CANP small subunit). | |||||
|
CPNS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.475407 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96L46, Q9BPV4 | Gene names | CAPNS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
CPNS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.472034 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D7J7 | Gene names | Capns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
GRAN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.362472 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P28676 | Gene names | GCA, GCL | |||
|
Domain Architecture |
|
|||||
| Description | Grancalcin. | |||||
|
GRAN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.370780 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VC88, Q8BL53, Q8K3Z6 | Gene names | Gca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grancalcin. | |||||
|
PDCD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.279165 (rank : 30) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75340 | Gene names | PDCD6, ALG2 | |||
|
Domain Architecture |
|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2). | |||||
|
PDCD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.277776 (rank : 31) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P12815, Q545I0, Q61145 | Gene names | Pdcd6, Alg2 | |||
|
Domain Architecture |
|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2) (PMP41) (ALG-257). | |||||
|
PEF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.332137 (rank : 29) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UBV8 | Gene names | PEF1, ABP32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
PEF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.341472 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BFY6, Q8VCT5, Q9CYW8, Q9D934 | Gene names | Pef1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
SORCN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.365087 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P30626 | Gene names | SRI | |||
|
Domain Architecture |
|
|||||
| Description | Sorcin (22 kDa protein) (CP-22) (V19). | |||||
|
SORCN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.368025 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P069, Q3UKC5, Q9CR38, Q9D7V8 | Gene names | Sri | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorcin. | |||||
|
TNNC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.050631 (rank : 36) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P19123 | Gene names | Tnnc1, Tncc | |||
|
Domain Architecture |
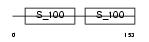
|
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
CAN6_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35646 | Gene names | Capn6, Capa6 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6. | |||||
|
CAN6_HUMAN
|
||||||
| NC score | 0.999578 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y6Q1, Q9UEQ1, Q9UJA8 | Gene names | CAPN6, CALPM, CANPX | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6 (Calpamodulin) (CalpM) (Calpain-like protease X-linked). | |||||
|
CAN5_HUMAN
|
||||||
| NC score | 0.987572 (rank : 3) | θ value | 3.47952e-169 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15484, O00263 | Gene names | CAPN5 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-5 (EC 3.4.22.-) (nCL-3) (htra-3). | |||||
|
CAN5_MOUSE
|
||||||
| NC score | 0.987233 (rank : 4) | θ value | 5.36813e-170 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08688, Q91YU0 | Gene names | Capn5 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-5 (EC 3.4.22.-) (nCL-3). | |||||
|
CAN10_MOUSE
|
||||||
| NC score | 0.918285 (rank : 5) | θ value | 3.48949e-44 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ESK3, Q99J13, Q9WVF0 | Gene names | Capn10, Capn8 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
CAN10_HUMAN
|
||||||
| NC score | 0.914336 (rank : 6) | θ value | 3.61106e-41 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HC96, Q8NCD4, Q96IG4, Q96JI2, Q9HC89, Q9HC90, Q9HC91, Q9HC92, Q9HC93, Q9HC94, Q9HC95 | Gene names | CAPN10, KIAA1845 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
CAN12_HUMAN
|
||||||
| NC score | 0.902450 (rank : 7) | θ value | 8.55514e-59 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZSI9 | Gene names | CAPN12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain-12 (EC 3.4.22.-). | |||||
|
CAN12_MOUSE
|
||||||
| NC score | 0.901330 (rank : 8) | θ value | 1.07972e-61 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ER56, Q9ER53, Q9ER54, Q9ER55 | Gene names | Capn12 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-12 (EC 3.4.22.-). | |||||
|
CAN9_MOUSE
|
||||||
| NC score | 0.884082 (rank : 9) | θ value | 7.19216e-82 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D805, O35919 | Gene names | Capn9, Ncl4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4). | |||||
|
CAN9_HUMAN
|
||||||
| NC score | 0.883968 (rank : 10) | θ value | 5.69867e-79 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14815, Q9NS74 | Gene names | CAPN9, NCL4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4) (CG36 protein). | |||||
|
CAN11_HUMAN
|
||||||
| NC score | 0.882701 (rank : 11) | θ value | 3.34864e-71 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UMQ6, Q8N4R5 | Gene names | CAPN11 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-11 (EC 3.4.22.-) (Calcium-activated neutral proteinase 11) (CANP 11). | |||||
|
CAN2_HUMAN
|
||||||
| NC score | 0.881548 (rank : 12) | θ value | 2.1655e-78 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17655, Q16738, Q6PJT3, Q8WU26, Q9HBB1 | Gene names | CAPN2, CANPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (Calpain large polypeptide L2). | |||||
|
CAN2_MOUSE
|
||||||
| NC score | 0.879035 (rank : 13) | θ value | 2.82823e-78 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08529, O35518, O54843 | Gene names | Capn2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (80 kDa M-calpain subunit) (CALP80). | |||||
|
CAN1_MOUSE
|
||||||
| NC score | 0.871100 (rank : 14) | θ value | 3.23591e-74 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35350, O88666 | Gene names | Capn1, Canp1, Capa1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
CAN1_HUMAN
|
||||||
| NC score | 0.869982 (rank : 15) | θ value | 1.66191e-70 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P07384 | Gene names | CAPN1, CANPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
CAN3_HUMAN
|
||||||
| NC score | 0.866187 (rank : 16) | θ value | 3.12697e-77 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20807, Q9BTU4, Q9Y5S6, Q9Y5S7 | Gene names | CAPN3, CANP3, CANPL3, NCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3) (nCL-1). | |||||
|
CAN3_MOUSE
|
||||||
| NC score | 0.862866 (rank : 17) | θ value | 1.26953e-78 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64691, Q9WUC5 | Gene names | Capn3 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3). | |||||
|
CAN7_MOUSE
|
||||||
| NC score | 0.751242 (rank : 18) | θ value | 2.43343e-21 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R1S8, Q9Z0P9 | Gene names | Capn7, Palbh | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
CAN7_HUMAN
|
||||||
| NC score | 0.750222 (rank : 19) | θ value | 1.86321e-21 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6W3 | Gene names | CAPN7, PALBH | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
CPNS1_MOUSE
|
||||||
| NC score | 0.482685 (rank : 20) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88456, Q3V0X5, Q5BKQ2, Q8CEI2, Q8VEK4, Q9R1C5 | Gene names | Capns1, Capn4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 1 (CSS1) (Calcium-dependent protease small subunit 1) (Calcium-dependent protease small subunit) (CDPS) (Calpain regulatory subunit) (Calcium-activated neutral proteinase small subunit) (CANP small subunit). | |||||
|
CPNS1_HUMAN
|
||||||
| NC score | 0.479219 (rank : 21) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P04632, Q8WTX3, Q96EW0 | Gene names | CAPNS1, CAPN4, CAPNS | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 1 (CSS1) (Calcium-dependent protease small subunit 1) (Calcium-dependent protease small subunit) (CDPS) (Calpain regulatory subunit) (Calcium-activated neutral proteinase small subunit) (CANP small subunit). | |||||
|
CPNS2_HUMAN
|
||||||
| NC score | 0.475407 (rank : 22) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96L46, Q9BPV4 | Gene names | CAPNS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
CPNS2_MOUSE
|
||||||
| NC score | 0.472034 (rank : 23) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D7J7 | Gene names | Capns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
GRAN_MOUSE
|
||||||
| NC score | 0.370780 (rank : 24) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VC88, Q8BL53, Q8K3Z6 | Gene names | Gca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grancalcin. | |||||
|
SORCN_MOUSE
|
||||||
| NC score | 0.368025 (rank : 25) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P069, Q3UKC5, Q9CR38, Q9D7V8 | Gene names | Sri | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorcin. | |||||
|
SORCN_HUMAN
|
||||||
| NC score | 0.365087 (rank : 26) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P30626 | Gene names | SRI | |||
|
Domain Architecture |

|
|||||
| Description | Sorcin (22 kDa protein) (CP-22) (V19). | |||||
|
GRAN_HUMAN
|
||||||
| NC score | 0.362472 (rank : 27) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P28676 | Gene names | GCA, GCL | |||
|
Domain Architecture |

|
|||||
| Description | Grancalcin. | |||||
|
PEF1_MOUSE
|
||||||
| NC score | 0.341472 (rank : 28) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BFY6, Q8VCT5, Q9CYW8, Q9D934 | Gene names | Pef1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
PEF1_HUMAN
|
||||||
| NC score | 0.332137 (rank : 29) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UBV8 | Gene names | PEF1, ABP32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
PDCD6_HUMAN
|
||||||
| NC score | 0.279165 (rank : 30) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75340 | Gene names | PDCD6, ALG2 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2). | |||||
|
PDCD6_MOUSE
|
||||||
| NC score | 0.277776 (rank : 31) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P12815, Q545I0, Q61145 | Gene names | Pdcd6, Alg2 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2) (PMP41) (ALG-257). | |||||
|
CF103_HUMAN
|
||||||
| NC score | 0.182148 (rank : 32) | θ value | 0.0330416 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N7X0, Q5T402 | Gene names | C6orf103 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf103. | |||||
|
CALL5_HUMAN
|
||||||
| NC score | 0.080753 (rank : 33) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
CALM4_MOUSE
|
||||||
| NC score | 0.061183 (rank : 34) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
RFIP2_HUMAN
|
||||||
| NC score | 0.051922 (rank : 35) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7L804, Q9Y2F0 | Gene names | RAB11FIP2, KIAA0941 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 2 (Rab11-FIP2) (NRip11). | |||||
|
TNNC1_MOUSE
|
||||||
| NC score | 0.050631 (rank : 36) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P19123 | Gene names | Tnnc1, Tncc | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
UN13A_HUMAN
|
||||||
| NC score | 0.043333 (rank : 37) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
UN13B_MOUSE
|
||||||
| NC score | 0.034819 (rank : 38) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z1N9 | Gene names | Unc13b, Unc13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
UN13B_HUMAN
|
||||||
| NC score | 0.029082 (rank : 39) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14795 | Gene names | UNC13B, UNC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
RFIP1_MOUSE
|
||||||
| NC score | 0.027524 (rank : 40) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D620, Q3UBC2, Q8BN24 | Gene names | Rab11fip1, Rcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
RFIP1_HUMAN
|
||||||
| NC score | 0.023191 (rank : 41) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
NIBA_HUMAN
|
||||||
| NC score | 0.013999 (rank : 42) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZQ8, Q5TEM8, Q8TEI5, Q9H593, Q9H9Y8, Q9HCB9 | Gene names | C1orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban protein. | |||||
|
ZN292_HUMAN
|
||||||
| NC score | 0.003224 (rank : 43) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||