Please be patient as the page loads
|
CAN9_MOUSE
|
||||||
| SwissProt Accessions | Q9D805, O35919 | Gene names | Capn9, Ncl4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CAN11_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.989979 (rank : 5) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UMQ6, Q8N4R5 | Gene names | CAPN11 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-11 (EC 3.4.22.-) (Calcium-activated neutral proteinase 11) (CANP 11). | |||||
|
CAN1_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.989769 (rank : 6) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P07384 | Gene names | CAPN1, CANPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
CAN1_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.989526 (rank : 7) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O35350, O88666 | Gene names | Capn1, Canp1, Capa1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
CAN2_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.991313 (rank : 4) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P17655, Q16738, Q6PJT3, Q8WU26, Q9HBB1 | Gene names | CAPN2, CANPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (Calpain large polypeptide L2). | |||||
|
CAN2_MOUSE
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.991403 (rank : 3) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O08529, O35518, O54843 | Gene names | Capn2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (80 kDa M-calpain subunit) (CALP80). | |||||
|
CAN3_HUMAN
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.988110 (rank : 8) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P20807, Q9BTU4, Q9Y5S6, Q9Y5S7 | Gene names | CAPN3, CANP3, CANPL3, NCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3) (nCL-1). | |||||
|
CAN3_MOUSE
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.985104 (rank : 9) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q64691, Q9WUC5 | Gene names | Capn3 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3). | |||||
|
CAN9_HUMAN
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.998042 (rank : 2) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O14815, Q9NS74 | Gene names | CAPN9, NCL4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4) (CG36 protein). | |||||
|
CAN9_MOUSE
|
||||||
| θ value | 0 (rank : 9) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9D805, O35919 | Gene names | Capn9, Ncl4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4). | |||||
|
CAN12_HUMAN
|
||||||
| θ value | 7.07634e-138 (rank : 10) | NC score | 0.978109 (rank : 10) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6ZSI9 | Gene names | CAPN12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain-12 (EC 3.4.22.-). | |||||
|
CAN12_MOUSE
|
||||||
| θ value | 3.51195e-137 (rank : 11) | NC score | 0.977672 (rank : 11) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9ER56, Q9ER53, Q9ER54, Q9ER55 | Gene names | Capn12 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-12 (EC 3.4.22.-). | |||||
|
CAN5_MOUSE
|
||||||
| θ value | 2.46047e-98 (rank : 12) | NC score | 0.897258 (rank : 12) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08688, Q91YU0 | Gene names | Capn5 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-5 (EC 3.4.22.-) (nCL-3). | |||||
|
CAN5_HUMAN
|
||||||
| θ value | 2.08291e-97 (rank : 13) | NC score | 0.896513 (rank : 13) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15484, O00263 | Gene names | CAPN5 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-5 (EC 3.4.22.-) (nCL-3) (htra-3). | |||||
|
CAN6_HUMAN
|
||||||
| θ value | 2.91998e-83 (rank : 14) | NC score | 0.885009 (rank : 14) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y6Q1, Q9UEQ1, Q9UJA8 | Gene names | CAPN6, CALPM, CANPX | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6 (Calpamodulin) (CalpM) (Calpain-like protease X-linked). | |||||
|
CAN6_MOUSE
|
||||||
| θ value | 7.19216e-82 (rank : 15) | NC score | 0.884082 (rank : 15) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35646 | Gene names | Capn6, Capa6 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6. | |||||
|
CAN10_MOUSE
|
||||||
| θ value | 2.10721e-57 (rank : 16) | NC score | 0.868742 (rank : 16) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ESK3, Q99J13, Q9WVF0 | Gene names | Capn10, Capn8 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
CAN10_HUMAN
|
||||||
| θ value | 1.72781e-51 (rank : 17) | NC score | 0.864346 (rank : 17) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HC96, Q8NCD4, Q96IG4, Q96JI2, Q9HC89, Q9HC90, Q9HC91, Q9HC92, Q9HC93, Q9HC94, Q9HC95 | Gene names | CAPN10, KIAA1845 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
CPNS1_MOUSE
|
||||||
| θ value | 8.04462e-41 (rank : 18) | NC score | 0.810138 (rank : 18) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O88456, Q3V0X5, Q5BKQ2, Q8CEI2, Q8VEK4, Q9R1C5 | Gene names | Capns1, Capn4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 1 (CSS1) (Calcium-dependent protease small subunit 1) (Calcium-dependent protease small subunit) (CDPS) (Calpain regulatory subunit) (Calcium-activated neutral proteinase small subunit) (CANP small subunit). | |||||
|
CPNS1_HUMAN
|
||||||
| θ value | 1.79215e-40 (rank : 19) | NC score | 0.806266 (rank : 20) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P04632, Q8WTX3, Q96EW0 | Gene names | CAPNS1, CAPN4, CAPNS | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 1 (CSS1) (Calcium-dependent protease small subunit 1) (Calcium-dependent protease small subunit) (CDPS) (Calpain regulatory subunit) (Calcium-activated neutral proteinase small subunit) (CANP small subunit). | |||||
|
CPNS2_HUMAN
|
||||||
| θ value | 1.85459e-37 (rank : 20) | NC score | 0.808443 (rank : 19) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96L46, Q9BPV4 | Gene names | CAPNS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
CPNS2_MOUSE
|
||||||
| θ value | 2.67802e-36 (rank : 21) | NC score | 0.802990 (rank : 21) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D7J7 | Gene names | Capns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
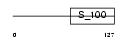
SORCN_HUMAN
|
||||||
| θ value | 5.99374e-20 (rank : 22) | NC score | 0.693157 (rank : 24) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P30626 | Gene names | SRI | |||
|
Domain Architecture |
|
|||||
| Description | Sorcin (22 kDa protein) (CP-22) (V19). | |||||
|
SORCN_MOUSE
|
||||||
| θ value | 1.74391e-19 (rank : 23) | NC score | 0.696961 (rank : 23) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6P069, Q3UKC5, Q9CR38, Q9D7V8 | Gene names | Sri | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorcin. | |||||
|
GRAN_MOUSE
|
||||||
| θ value | 5.07402e-19 (rank : 24) | NC score | 0.698251 (rank : 22) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8VC88, Q8BL53, Q8K3Z6 | Gene names | Gca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grancalcin. | |||||
|
CAN7_MOUSE
|
||||||
| θ value | 4.29542e-18 (rank : 25) | NC score | 0.671614 (rank : 26) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R1S8, Q9Z0P9 | Gene names | Capn7, Palbh | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
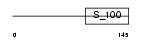
GRAN_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 26) | NC score | 0.689125 (rank : 25) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P28676 | Gene names | GCA, GCL | |||
|
Domain Architecture |
|
|||||
| Description | Grancalcin. | |||||
|
CAN7_HUMAN
|
||||||
| θ value | 9.56915e-18 (rank : 27) | NC score | 0.669208 (rank : 27) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y6W3 | Gene names | CAPN7, PALBH | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
PEF1_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 28) | NC score | 0.648354 (rank : 28) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BFY6, Q8VCT5, Q9CYW8, Q9D934 | Gene names | Pef1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
PEF1_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 29) | NC score | 0.633506 (rank : 29) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UBV8 | Gene names | PEF1, ABP32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
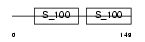
PDCD6_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 30) | NC score | 0.580670 (rank : 30) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75340 | Gene names | PDCD6, ALG2 | |||
|
Domain Architecture |
|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2). | |||||
|
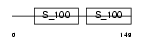
PDCD6_MOUSE
|
||||||
| θ value | 8.67504e-11 (rank : 31) | NC score | 0.578880 (rank : 31) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P12815, Q545I0, Q61145 | Gene names | Pdcd6, Alg2 | |||
|
Domain Architecture |
|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2) (PMP41) (ALG-257). | |||||
|
CALL5_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 32) | NC score | 0.204533 (rank : 32) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
CALM4_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 33) | NC score | 0.167400 (rank : 33) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
TNNC1_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 34) | NC score | 0.129243 (rank : 38) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P63316, O14800, P02590, P04463 | Gene names | TNNC1, TNNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
TNNC1_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 35) | NC score | 0.135062 (rank : 35) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P19123 | Gene names | Tnnc1, Tncc | |||
|
Domain Architecture |
|
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
CABP7_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 36) | NC score | 0.105309 (rank : 47) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86V35 | Gene names | CABP7 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CABP7_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 37) | NC score | 0.105309 (rank : 48) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91ZM8 | Gene names | Cabp7 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
PLSI_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 38) | NC score | 0.100771 (rank : 51) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14651 | Gene names | PLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-1 (I-plastin) (Intestine-specific plastin). | |||||
|
CANB1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 39) | NC score | 0.102836 (rank : 49) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P63098, P06705, P15117, Q08044 | Gene names | PPP3R1, CNA2, CNB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 40) | NC score | 0.102814 (rank : 50) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q63810 | Gene names | Ppp3r1, Cnb | |||
|
Domain Architecture |
|
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 41) | NC score | 0.090018 (rank : 60) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96LZ3, Q7Z4V8, Q8WYJ4 | Gene names | PPP3R2, CBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform) (Calcineurin B-like protein) (CBLP) (CNBII). | |||||
|
CALL3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 42) | NC score | 0.127968 (rank : 39) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
CABP4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 43) | NC score | 0.124472 (rank : 41) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CABP2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 44) | NC score | 0.094753 (rank : 55) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NPB3 | Gene names | CABP2 | |||
|
Domain Architecture |
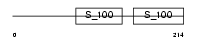
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CABP2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 45) | NC score | 0.097524 (rank : 52) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JLK4, Q9JLK5 | Gene names | Cabp2 | |||
|
Domain Architecture |
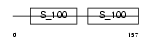
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CANB2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 46) | NC score | 0.090641 (rank : 59) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q63811 | Gene names | Ppp3r2 | |||
|
Domain Architecture |
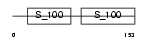
|
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform). | |||||
|
S100G_MOUSE
|
||||||
| θ value | 0.279714 (rank : 47) | NC score | 0.038394 (rank : 78) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97816 | Gene names | S100g, Calb3, S100d | |||
|
Domain Architecture |
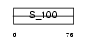
|
|||||
| Description | Protein S100-G (S100 calcium-binding protein G) (Vitamin D-dependent calcium-binding protein, intestinal) (CABP) (Calbindin D9K). | |||||
|
DMD_MOUSE
|
||||||
| θ value | 0.365318 (rank : 48) | NC score | 0.005271 (rank : 86) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
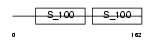
CETN1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 49) | NC score | 0.136911 (rank : 34) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q12798 | Gene names | CETN1, CEN1, CETN | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-1 (Caltractin isoform 2). | |||||
|
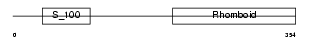
RHBD4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.039266 (rank : 77) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P58872 | Gene names | RHBDL4, VRHO | |||
|
Domain Architecture |
|
|||||
| Description | Rhomboid-related protein 4 (EC 3.4.21.105) (Ventrhoid transmembrane protein). | |||||
|
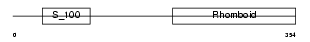
RHBD4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.037303 (rank : 79) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P58873 | Gene names | Rhbdl4, Vrho | |||
|
Domain Architecture |
|
|||||
| Description | Rhomboid-related protein 4 (EC 3.4.21.105) (Ventrhoid transmembrane protein). | |||||
|
CSEN_MOUSE
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.045502 (rank : 75) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QXT8, Q924L0, Q99PH9, Q99PI0, Q99PI2, Q99PI3, Q9JHZ5 | Gene names | Kcnip3, Csen, Dream, Kchip3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (A-type potassium channel modulatory protein 3) (KChIP3). | |||||
|
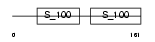
CABP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.096662 (rank : 54) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NZU7, O95663, Q9NZU8 | Gene names | CABP1 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 1 (CaBP1) (Calbrain). | |||||
|
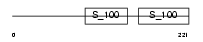
CABP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.097224 (rank : 53) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JLK7, Q9JLK6 | Gene names | Cabp1 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 1 (CaBP1). | |||||
|
RECQ4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.008122 (rank : 84) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
SPTA2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.024815 (rank : 82) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
TNNC2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.113017 (rank : 44) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P20801 | Gene names | Tnnc2, Tncs | |||
|
Domain Architecture |
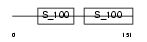
|
|||||
| Description | Troponin C, skeletal muscle (STNC). | |||||
|
CETN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.116705 (rank : 43) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
TNNC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.109591 (rank : 46) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P02585 | Gene names | TNNC2 | |||
|
Domain Architecture |
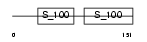
|
|||||
| Description | Troponin C, skeletal muscle. | |||||
|
CSEN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.040736 (rank : 76) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y2W7, Q53TJ5, Q96T40, Q9UJ84, Q9UJ85 | Gene names | KCNIP3, CSEN, DREAM, KCHIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (KChIP3) (A-type potassium channel modulatory protein 3). | |||||
|
FAS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.006980 (rank : 85) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49327, Q13479, Q16702, Q6P4U5, Q6SS02, Q969R1, Q96C68, Q96IT0 | Gene names | FASN, FAS | |||
|
Domain Architecture |

|
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
PLST_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.034151 (rank : 80) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P13797, Q86YI6 | Gene names | PLS3 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-3 (T-plastin). | |||||
|
CCD39_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.000890 (rank : 90) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CETN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.123135 (rank : 42) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P41209, Q3V119, Q9D9G9, Q9DAL6 | Gene names | Cetn1, Calt | |||
|
Domain Architecture |
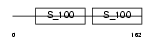
|
|||||
| Description | Centrin-1 (Caltractin). | |||||
|
DUOX2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.029523 (rank : 81) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NRD8, Q9NR02, Q9UHF9 | Gene names | DUOX2, LNOX2, THOX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 2 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH oxidase/peroxidase DUOX2) (NADPH thyroid oxidase 2) (Thyroid oxidase 2) (NADH/NADPH thyroid oxidase p138-tox) (p138 thyroid oxidase) (Large NOX 2) (Long NOX 2). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | -0.000561 (rank : 91) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
CETN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.112067 (rank : 45) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P41208, Q53XW1 | Gene names | CETN2, CALT, CEN2 | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
EDG6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.011595 (rank : 83) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 462 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0L1, Q3U4H3, Q8C6E2, Q8K1R9 | Gene names | Edg6, Lpc1, S1p4 | |||
|
Domain Architecture |
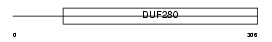
|
|||||
| Description | Sphingosine 1-phosphate receptor Edg-6 (S1P receptor Edg-6) (Endothelial differentiation G-protein coupled receptor 6) (Lysophospholipid receptor C1) (Sphingosine 1-phosphate receptor 4) (S1P4). | |||||
|
MOR2B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.004058 (rank : 88) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C5W4, Q8CG24 | Gene names | Morc2b, Tce6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 2B (TCE6). | |||||
|
CABP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.092069 (rank : 58) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CALB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.088171 (rank : 62) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
EFHC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.004646 (rank : 87) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D485, Q0VBF5, Q8CDV9 | Gene names | Efhc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member C2. | |||||
|
UTRO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.003421 (rank : 89) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
CABP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.073801 (rank : 67) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZU6, Q9NYY0, Q9NZT9 | Gene names | CABP3 | |||
|
Domain Architecture |
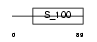
|
|||||
| Description | Calcium-binding protein 3 (CaBP3). | |||||
|
CABP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.088296 (rank : 61) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CABP5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.075131 (rank : 66) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CALB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.077822 (rank : 65) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P05937 | Gene names | CALB1, CAB27 | |||
|
Domain Architecture |
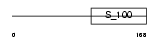
|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K). | |||||
|
CALB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.082179 (rank : 63) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P12658, Q545N6 | Gene names | Calb1 | |||
|
Domain Architecture |
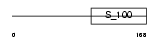
|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K) (Spot 35 protein) (PCD-29). | |||||
|
CALB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.061031 (rank : 73) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
CALL6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.078766 (rank : 64) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8TD86, Q6Q2C4 | Gene names | CALML6, CAGLP, CALGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-like protein 6 (Calglandulin-like protein). | |||||
|
CALM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.132265 (rank : 36) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P62158, P02593, P70667, P99014, Q13942, Q53S29, Q61379, Q61380, Q96HK3 | Gene names | CALM1, CALM, CAM, CAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.132265 (rank : 37) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P62204, P02593, P70667, P99014, Q61379, Q61380 | Gene names | Calm1, Calm, Cam, Cam1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.068127 (rank : 69) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BXU9 | Gene names | CALN1, CABP8 | |||
|
Domain Architecture |
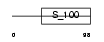
|
|||||
| Description | Calneuron-1 (Calcium-binding protein CaBP8). | |||||
|
CALN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.068047 (rank : 70) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JJG7 | Gene names | Caln1 | |||
|
Domain Architecture |
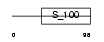
|
|||||
| Description | Calneuron-1. | |||||
|
CAYP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.069950 (rank : 68) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13938, Q3MJA7, Q8WUZ3 | Gene names | CAPS | |||
|
Domain Architecture |
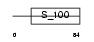
|
|||||
| Description | Calcyphosin (Calcyphosine). | |||||
|
CETN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.092576 (rank : 57) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O15182, Q9BS23 | Gene names | CETN3, CEN3 | |||
|
Domain Architecture |
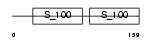
|
|||||
| Description | Centrin-3. | |||||
|
CETN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.092871 (rank : 56) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35648 | Gene names | Cetn3, Cen3 | |||
|
Domain Architecture |
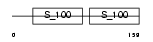
|
|||||
| Description | Centrin-3. | |||||
|
CF103_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.125604 (rank : 40) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N7X0, Q5T402 | Gene names | C6orf103 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf103. | |||||
|
PRVA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.061809 (rank : 72) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P32848 | Gene names | Pvalb, Pva | |||
|
Domain Architecture |
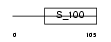
|
|||||
| Description | Parvalbumin alpha. | |||||
|
SEGN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.066935 (rank : 71) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O76038, Q5VV44, Q96QV7, Q9UJF6 | Gene names | SCGN, SECRET | |||
|
Domain Architecture |
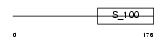
|
|||||
| Description | Secretagogin. | |||||
|
SEGN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.057908 (rank : 74) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91WD9 | Gene names | Scgn | |||
|
Domain Architecture |

|
|||||
| Description | Secretagogin. | |||||
|
CAN9_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 9) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9D805, O35919 | Gene names | Capn9, Ncl4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4). | |||||
|
CAN9_HUMAN
|
||||||
| NC score | 0.998042 (rank : 2) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O14815, Q9NS74 | Gene names | CAPN9, NCL4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4) (CG36 protein). | |||||
|
CAN2_MOUSE
|
||||||
| NC score | 0.991403 (rank : 3) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O08529, O35518, O54843 | Gene names | Capn2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (80 kDa M-calpain subunit) (CALP80). | |||||
|
CAN2_HUMAN
|
||||||
| NC score | 0.991313 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P17655, Q16738, Q6PJT3, Q8WU26, Q9HBB1 | Gene names | CAPN2, CANPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (Calpain large polypeptide L2). | |||||
|
CAN11_HUMAN
|
||||||
| NC score | 0.989979 (rank : 5) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UMQ6, Q8N4R5 | Gene names | CAPN11 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-11 (EC 3.4.22.-) (Calcium-activated neutral proteinase 11) (CANP 11). | |||||
|
CAN1_HUMAN
|
||||||
| NC score | 0.989769 (rank : 6) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P07384 | Gene names | CAPN1, CANPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
CAN1_MOUSE
|
||||||
| NC score | 0.989526 (rank : 7) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O35350, O88666 | Gene names | Capn1, Canp1, Capa1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
CAN3_HUMAN
|
||||||
| NC score | 0.988110 (rank : 8) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P20807, Q9BTU4, Q9Y5S6, Q9Y5S7 | Gene names | CAPN3, CANP3, CANPL3, NCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3) (nCL-1). | |||||
|
CAN3_MOUSE
|
||||||
| NC score | 0.985104 (rank : 9) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q64691, Q9WUC5 | Gene names | Capn3 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3). | |||||
|
CAN12_HUMAN
|
||||||
| NC score | 0.978109 (rank : 10) | θ value | 7.07634e-138 (rank : 10) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6ZSI9 | Gene names | CAPN12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain-12 (EC 3.4.22.-). | |||||
|
CAN12_MOUSE
|
||||||
| NC score | 0.977672 (rank : 11) | θ value | 3.51195e-137 (rank : 11) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9ER56, Q9ER53, Q9ER54, Q9ER55 | Gene names | Capn12 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-12 (EC 3.4.22.-). | |||||
|
CAN5_MOUSE
|
||||||
| NC score | 0.897258 (rank : 12) | θ value | 2.46047e-98 (rank : 12) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08688, Q91YU0 | Gene names | Capn5 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-5 (EC 3.4.22.-) (nCL-3). | |||||
|
CAN5_HUMAN
|
||||||
| NC score | 0.896513 (rank : 13) | θ value | 2.08291e-97 (rank : 13) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15484, O00263 | Gene names | CAPN5 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-5 (EC 3.4.22.-) (nCL-3) (htra-3). | |||||
|
CAN6_HUMAN
|
||||||
| NC score | 0.885009 (rank : 14) | θ value | 2.91998e-83 (rank : 14) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y6Q1, Q9UEQ1, Q9UJA8 | Gene names | CAPN6, CALPM, CANPX | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6 (Calpamodulin) (CalpM) (Calpain-like protease X-linked). | |||||
|
CAN6_MOUSE
|
||||||
| NC score | 0.884082 (rank : 15) | θ value | 7.19216e-82 (rank : 15) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35646 | Gene names | Capn6, Capa6 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6. | |||||
|
CAN10_MOUSE
|
||||||
| NC score | 0.868742 (rank : 16) | θ value | 2.10721e-57 (rank : 16) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ESK3, Q99J13, Q9WVF0 | Gene names | Capn10, Capn8 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
CAN10_HUMAN
|
||||||
| NC score | 0.864346 (rank : 17) | θ value | 1.72781e-51 (rank : 17) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HC96, Q8NCD4, Q96IG4, Q96JI2, Q9HC89, Q9HC90, Q9HC91, Q9HC92, Q9HC93, Q9HC94, Q9HC95 | Gene names | CAPN10, KIAA1845 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
CPNS1_MOUSE
|
||||||
| NC score | 0.810138 (rank : 18) | θ value | 8.04462e-41 (rank : 18) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O88456, Q3V0X5, Q5BKQ2, Q8CEI2, Q8VEK4, Q9R1C5 | Gene names | Capns1, Capn4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 1 (CSS1) (Calcium-dependent protease small subunit 1) (Calcium-dependent protease small subunit) (CDPS) (Calpain regulatory subunit) (Calcium-activated neutral proteinase small subunit) (CANP small subunit). | |||||
|
CPNS2_HUMAN
|
||||||
| NC score | 0.808443 (rank : 19) | θ value | 1.85459e-37 (rank : 20) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96L46, Q9BPV4 | Gene names | CAPNS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
CPNS1_HUMAN
|
||||||
| NC score | 0.806266 (rank : 20) | θ value | 1.79215e-40 (rank : 19) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P04632, Q8WTX3, Q96EW0 | Gene names | CAPNS1, CAPN4, CAPNS | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 1 (CSS1) (Calcium-dependent protease small subunit 1) (Calcium-dependent protease small subunit) (CDPS) (Calpain regulatory subunit) (Calcium-activated neutral proteinase small subunit) (CANP small subunit). | |||||
|
CPNS2_MOUSE
|
||||||
| NC score | 0.802990 (rank : 21) | θ value | 2.67802e-36 (rank : 21) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D7J7 | Gene names | Capns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
GRAN_MOUSE
|
||||||
| NC score | 0.698251 (rank : 22) | θ value | 5.07402e-19 (rank : 24) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8VC88, Q8BL53, Q8K3Z6 | Gene names | Gca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grancalcin. | |||||
|
SORCN_MOUSE
|
||||||
| NC score | 0.696961 (rank : 23) | θ value | 1.74391e-19 (rank : 23) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6P069, Q3UKC5, Q9CR38, Q9D7V8 | Gene names | Sri | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorcin. | |||||
|
SORCN_HUMAN
|
||||||
| NC score | 0.693157 (rank : 24) | θ value | 5.99374e-20 (rank : 22) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P30626 | Gene names | SRI | |||
|
Domain Architecture |

|
|||||
| Description | Sorcin (22 kDa protein) (CP-22) (V19). | |||||
|
GRAN_HUMAN
|
||||||
| NC score | 0.689125 (rank : 25) | θ value | 7.32683e-18 (rank : 26) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P28676 | Gene names | GCA, GCL | |||
|
Domain Architecture |

|
|||||
| Description | Grancalcin. | |||||
|
CAN7_MOUSE
|
||||||
| NC score | 0.671614 (rank : 26) | θ value | 4.29542e-18 (rank : 25) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R1S8, Q9Z0P9 | Gene names | Capn7, Palbh | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
CAN7_HUMAN
|
||||||
| NC score | 0.669208 (rank : 27) | θ value | 9.56915e-18 (rank : 27) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y6W3 | Gene names | CAPN7, PALBH | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
PEF1_MOUSE
|
||||||
| NC score | 0.648354 (rank : 28) | θ value | 4.59992e-12 (rank : 28) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BFY6, Q8VCT5, Q9CYW8, Q9D934 | Gene names | Pef1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
PEF1_HUMAN
|
||||||
| NC score | 0.633506 (rank : 29) | θ value | 2.28291e-11 (rank : 29) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UBV8 | Gene names | PEF1, ABP32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
PDCD6_HUMAN
|
||||||
| NC score | 0.580670 (rank : 30) | θ value | 8.67504e-11 (rank : 30) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75340 | Gene names | PDCD6, ALG2 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2). | |||||
|
PDCD6_MOUSE
|
||||||
| NC score | 0.578880 (rank : 31) | θ value | 8.67504e-11 (rank : 31) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P12815, Q545I0, Q61145 | Gene names | Pdcd6, Alg2 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2) (PMP41) (ALG-257). | |||||
|
CALL5_HUMAN
|
||||||
| NC score | 0.204533 (rank : 32) | θ value | 5.44631e-05 (rank : 32) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
CALM4_MOUSE
|
||||||
| NC score | 0.167400 (rank : 33) | θ value | 0.00298849 (rank : 33) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
CETN1_HUMAN
|
||||||
| NC score | 0.136911 (rank : 34) | θ value | 0.47712 (rank : 49) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q12798 | Gene names | CETN1, CEN1, CETN | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-1 (Caltractin isoform 2). | |||||
|
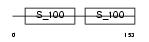
TNNC1_MOUSE
|
||||||
| NC score | 0.135062 (rank : 35) | θ value | 0.00298849 (rank : 35) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P19123 | Gene names | Tnnc1, Tncc | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
CALM_HUMAN
|
||||||
| NC score | 0.132265 (rank : 36) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P62158, P02593, P70667, P99014, Q13942, Q53S29, Q61379, Q61380, Q96HK3 | Gene names | CALM1, CALM, CAM, CAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALM_MOUSE
|
||||||
| NC score | 0.132265 (rank : 37) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P62204, P02593, P70667, P99014, Q61379, Q61380 | Gene names | Calm1, Calm, Cam, Cam1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
TNNC1_HUMAN
|
||||||
| NC score | 0.129243 (rank : 38) | θ value | 0.00298849 (rank : 34) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P63316, O14800, P02590, P04463 | Gene names | TNNC1, TNNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
CALL3_HUMAN
|
||||||
| NC score | 0.127968 (rank : 39) | θ value | 0.0563607 (rank : 42) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
CF103_HUMAN
|
||||||
| NC score | 0.125604 (rank : 40) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N7X0, Q5T402 | Gene names | C6orf103 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf103. | |||||
|
CABP4_HUMAN
|
||||||
| NC score | 0.124472 (rank : 41) | θ value | 0.0961366 (rank : 43) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CETN1_MOUSE
|
||||||
| NC score | 0.123135 (rank : 42) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P41209, Q3V119, Q9D9G9, Q9DAL6 | Gene names | Cetn1, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-1 (Caltractin). | |||||
|
CETN2_MOUSE
|
||||||
| NC score | 0.116705 (rank : 43) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
TNNC2_MOUSE
|
||||||
| NC score | 0.113017 (rank : 44) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P20801 | Gene names | Tnnc2, Tncs | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, skeletal muscle (STNC). | |||||
|
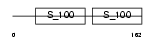
CETN2_HUMAN
|
||||||
| NC score | 0.112067 (rank : 45) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P41208, Q53XW1 | Gene names | CETN2, CALT, CEN2 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
TNNC2_HUMAN
|
||||||
| NC score | 0.109591 (rank : 46) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P02585 | Gene names | TNNC2 | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, skeletal muscle. | |||||
|
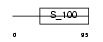
CABP7_HUMAN
|
||||||
| NC score | 0.105309 (rank : 47) | θ value | 0.0148317 (rank : 36) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86V35 | Gene names | CABP7 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
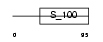
CABP7_MOUSE
|
||||||
| NC score | 0.105309 (rank : 48) | θ value | 0.0148317 (rank : 37) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91ZM8 | Gene names | Cabp7 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CANB1_HUMAN
|
||||||
| NC score | 0.102836 (rank : 49) | θ value | 0.0193708 (rank : 39) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P63098, P06705, P15117, Q08044 | Gene names | PPP3R1, CNA2, CNB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
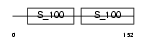
CANB1_MOUSE
|
||||||
| NC score | 0.102814 (rank : 50) | θ value | 0.0193708 (rank : 40) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q63810 | Gene names | Ppp3r1, Cnb | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
PLSI_HUMAN
|
||||||
| NC score | 0.100771 (rank : 51) | θ value | 0.0148317 (rank : 38) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14651 | Gene names | PLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-1 (I-plastin) (Intestine-specific plastin). | |||||
|
CABP2_MOUSE
|
||||||
| NC score | 0.097524 (rank : 52) | θ value | 0.279714 (rank : 45) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JLK4, Q9JLK5 | Gene names | Cabp2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CABP1_MOUSE
|
||||||
| NC score | 0.097224 (rank : 53) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JLK7, Q9JLK6 | Gene names | Cabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 1 (CaBP1). | |||||
|
CABP1_HUMAN
|
||||||
| NC score | 0.096662 (rank : 54) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NZU7, O95663, Q9NZU8 | Gene names | CABP1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 1 (CaBP1) (Calbrain). | |||||
|
CABP2_HUMAN
|
||||||
| NC score | 0.094753 (rank : 55) | θ value | 0.279714 (rank : 44) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NPB3 | Gene names | CABP2 | |||
|
Domain Architecture |
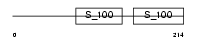
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CETN3_MOUSE
|
||||||
| NC score | 0.092871 (rank : 56) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35648 | Gene names | Cetn3, Cen3 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-3. | |||||
|
CETN3_HUMAN
|
||||||
| NC score | 0.092576 (rank : 57) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O15182, Q9BS23 | Gene names | CETN3, CEN3 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-3. | |||||
|
CABP4_MOUSE
|
||||||
| NC score | 0.092069 (rank : 58) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CANB2_MOUSE
|
||||||
| NC score | 0.090641 (rank : 59) | θ value | 0.279714 (rank : 46) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q63811 | Gene names | Ppp3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform). | |||||
|
CANB2_HUMAN
|
||||||
| NC score | 0.090018 (rank : 60) | θ value | 0.0431538 (rank : 41) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96LZ3, Q7Z4V8, Q8WYJ4 | Gene names | PPP3R2, CBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform) (Calcineurin B-like protein) (CBLP) (CNBII). | |||||
|
CABP5_HUMAN
|
||||||
| NC score | 0.088296 (rank : 61) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CALB2_HUMAN
|
||||||
| NC score | 0.088171 (rank : 62) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
CALB1_MOUSE
|
||||||
| NC score | 0.082179 (rank : 63) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P12658, Q545N6 | Gene names | Calb1 | |||
|
Domain Architecture |

|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K) (Spot 35 protein) (PCD-29). | |||||
|
CALL6_HUMAN
|
||||||
| NC score | 0.078766 (rank : 64) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8TD86, Q6Q2C4 | Gene names | CALML6, CAGLP, CALGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-like protein 6 (Calglandulin-like protein). | |||||
|
CALB1_HUMAN
|
||||||
| NC score | 0.077822 (rank : 65) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P05937 | Gene names | CALB1, CAB27 | |||
|
Domain Architecture |

|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K). | |||||
|
CABP5_MOUSE
|
||||||
| NC score | 0.075131 (rank : 66) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CABP3_HUMAN
|
||||||
| NC score | 0.073801 (rank : 67) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZU6, Q9NYY0, Q9NZT9 | Gene names | CABP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 3 (CaBP3). | |||||
|
CAYP1_HUMAN
|
||||||
| NC score | 0.069950 (rank : 68) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13938, Q3MJA7, Q8WUZ3 | Gene names | CAPS | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin (Calcyphosine). | |||||
|
CALN1_HUMAN
|
||||||
| NC score | 0.068127 (rank : 69) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BXU9 | Gene names | CALN1, CABP8 | |||
|
Domain Architecture |

|
|||||
| Description | Calneuron-1 (Calcium-binding protein CaBP8). | |||||
|
CALN1_MOUSE
|
||||||
| NC score | 0.068047 (rank : 70) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JJG7 | Gene names | Caln1 | |||
|
Domain Architecture |

|
|||||
| Description | Calneuron-1. | |||||
|
SEGN_HUMAN
|
||||||
| NC score | 0.066935 (rank : 71) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O76038, Q5VV44, Q96QV7, Q9UJF6 | Gene names | SCGN, SECRET | |||
|
Domain Architecture |

|
|||||
| Description | Secretagogin. | |||||
|
PRVA_MOUSE
|
||||||
| NC score | 0.061809 (rank : 72) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P32848 | Gene names | Pvalb, Pva | |||
|
Domain Architecture |

|
|||||
| Description | Parvalbumin alpha. | |||||
|
CALB2_MOUSE
|
||||||
| NC score | 0.061031 (rank : 73) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
SEGN_MOUSE
|
||||||
| NC score | 0.057908 (rank : 74) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91WD9 | Gene names | Scgn | |||
|
Domain Architecture |

|
|||||
| Description | Secretagogin. | |||||
|
CSEN_MOUSE
|
||||||
| NC score | 0.045502 (rank : 75) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QXT8, Q924L0, Q99PH9, Q99PI0, Q99PI2, Q99PI3, Q9JHZ5 | Gene names | Kcnip3, Csen, Dream, Kchip3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (A-type potassium channel modulatory protein 3) (KChIP3). | |||||
|
CSEN_HUMAN
|
||||||
| NC score | 0.040736 (rank : 76) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y2W7, Q53TJ5, Q96T40, Q9UJ84, Q9UJ85 | Gene names | KCNIP3, CSEN, DREAM, KCHIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (KChIP3) (A-type potassium channel modulatory protein 3). | |||||
|
RHBD4_HUMAN
|
||||||
| NC score | 0.039266 (rank : 77) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P58872 | Gene names | RHBDL4, VRHO | |||
|
Domain Architecture |

|
|||||
| Description | Rhomboid-related protein 4 (EC 3.4.21.105) (Ventrhoid transmembrane protein). | |||||
|
S100G_MOUSE
|
||||||
| NC score | 0.038394 (rank : 78) | θ value | 0.279714 (rank : 47) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97816 | Gene names | S100g, Calb3, S100d | |||
|
Domain Architecture |
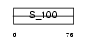
|
|||||
| Description | Protein S100-G (S100 calcium-binding protein G) (Vitamin D-dependent calcium-binding protein, intestinal) (CABP) (Calbindin D9K). | |||||
|
RHBD4_MOUSE
|
||||||
| NC score | 0.037303 (rank : 79) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P58873 | Gene names | Rhbdl4, Vrho | |||
|
Domain Architecture |

|
|||||
| Description | Rhomboid-related protein 4 (EC 3.4.21.105) (Ventrhoid transmembrane protein). | |||||
|
PLST_HUMAN
|
||||||
| NC score | 0.034151 (rank : 80) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P13797, Q86YI6 | Gene names | PLS3 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-3 (T-plastin). | |||||
|
DUOX2_HUMAN
|
||||||
| NC score | 0.029523 (rank : 81) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NRD8, Q9NR02, Q9UHF9 | Gene names | DUOX2, LNOX2, THOX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 2 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH oxidase/peroxidase DUOX2) (NADPH thyroid oxidase 2) (Thyroid oxidase 2) (NADH/NADPH thyroid oxidase p138-tox) (p138 thyroid oxidase) (Large NOX 2) (Long NOX 2). | |||||
|
SPTA2_HUMAN
|
||||||
| NC score | 0.024815 (rank : 82) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
EDG6_MOUSE
|
||||||
| NC score | 0.011595 (rank : 83) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 462 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0L1, Q3U4H3, Q8C6E2, Q8K1R9 | Gene names | Edg6, Lpc1, S1p4 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine 1-phosphate receptor Edg-6 (S1P receptor Edg-6) (Endothelial differentiation G-protein coupled receptor 6) (Lysophospholipid receptor C1) (Sphingosine 1-phosphate receptor 4) (S1P4). | |||||
|
RECQ4_HUMAN
|
||||||
| NC score | 0.008122 (rank : 84) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
FAS_HUMAN
|
||||||
| NC score | 0.006980 (rank : 85) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49327, Q13479, Q16702, Q6P4U5, Q6SS02, Q969R1, Q96C68, Q96IT0 | Gene names | FASN, FAS | |||
|
Domain Architecture |

|
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.005271 (rank : 86) | θ value | 0.365318 (rank : 48) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
EFHC2_MOUSE
|
||||||
| NC score | 0.004646 (rank : 87) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D485, Q0VBF5, Q8CDV9 | Gene names | Efhc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member C2. | |||||
|
MOR2B_MOUSE
|
||||||
| NC score | 0.004058 (rank : 88) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C5W4, Q8CG24 | Gene names | Morc2b, Tce6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 2B (TCE6). | |||||
|
UTRO_HUMAN
|
||||||
| NC score | 0.003421 (rank : 89) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
CCD39_HUMAN
|
||||||
| NC score | 0.000890 (rank : 90) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
HMMR_HUMAN
|
||||||
| NC score | -0.000561 (rank : 91) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||