Please be patient as the page loads
|
HM20B_MOUSE
|
||||||
| SwissProt Accessions | Q9Z104, Q9NSF7, Q9NSF8 | Gene names | Hmg20b, Braf35, Hmgx2, Smarce1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HM20B_MOUSE
|
||||||
| θ value | 4.75757e-126 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | Q9Z104, Q9NSF7, Q9NSF8 | Gene names | Hmg20b, Braf35, Hmgx2, Smarce1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35). | |||||
|
HM20B_HUMAN
|
||||||
| θ value | 1.17455e-116 (rank : 2) | NC score | 0.994391 (rank : 2) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q9P0W2, Q6IBP8, Q8NBD5, Q9HD21, Q9Y491, Q9Y4A2 | Gene names | HMG20B, BRAF35, HMGX2, SMARCE1R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35) (Sox-like transcriptional factor). | |||||
|
HM20A_HUMAN
|
||||||
| θ value | 2.09745e-73 (rank : 3) | NC score | 0.889140 (rank : 3) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9NP66, Q53G31, Q9NSF6 | Gene names | HMG20A, HMGX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (HMG domain-containing protein HMGX1). | |||||
|
HM20A_MOUSE
|
||||||
| θ value | 7.97034e-73 (rank : 4) | NC score | 0.886388 (rank : 4) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9DC33, Q3LSF9, Q6NV87, Q8BSK1, Q8C3C1, Q8CAA0, Q9CYG2 | Gene names | Hmg20a, Ibraf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (iBRAF) (Inhibitor of BRAF35) (HMG domain protein HMGX1). | |||||
|
HMGB3_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 5) | NC score | 0.707777 (rank : 6) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O15347, O95556, Q6NS40 | Gene names | HMGB3, HMG2A, HMG4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
HMGB3_MOUSE
|
||||||
| θ value | 7.34386e-10 (rank : 6) | NC score | 0.708126 (rank : 5) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O54879 | Gene names | Hmgb3, Hmg2a, Hmg4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
HMGB1_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 7) | NC score | 0.685974 (rank : 10) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P09429, Q6IBE1 | Gene names | HMGB1, HMG1 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HMGB1_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 8) | NC score | 0.686531 (rank : 9) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P63158, P07155, P27109, P27428 | Gene names | Hmgb1, Hmg-1, Hmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
SMCE1_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 9) | NC score | 0.578669 (rank : 20) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
SMCE1_MOUSE
|
||||||
| θ value | 1.63604e-09 (rank : 10) | NC score | 0.546197 (rank : 42) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
CN092_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 11) | NC score | 0.571015 (rank : 23) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
HMGB2_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 12) | NC score | 0.695659 (rank : 7) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P26583 | Gene names | HMGB2, HMG2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
SP100_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 13) | NC score | 0.510639 (rank : 56) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P23497, O75450, Q13343, Q96F70, Q96T24, Q96T95, Q9NP33, Q9UE32 | Gene names | SP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein) (Lysp100b). | |||||
|
TOX_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 14) | NC score | 0.575327 (rank : 21) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q66JW3, Q8BKH9, Q8BYQ5, Q8R4H0 | Gene names | Tox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
CN092_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 15) | NC score | 0.581294 (rank : 18) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O94842 | Gene names | C14orf92, KIAA0737 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
TOX_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 16) | NC score | 0.572948 (rank : 22) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O94900, Q96AV5 | Gene names | TOX, KIAA0808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
HMGB2_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 17) | NC score | 0.690357 (rank : 8) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P30681, Q3UXT1, Q9EQD5 | Gene names | Hmgb2, Hmg2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
HMG1X_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 18) | NC score | 0.657647 (rank : 11) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UGV6 | Gene names | HMG1L10 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 1-like 10 (HMG-1L10). | |||||
|
PB1_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 19) | NC score | 0.397584 (rank : 73) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
SSRP1_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 20) | NC score | 0.642269 (rank : 12) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
SSRP1_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 21) | NC score | 0.641945 (rank : 13) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 22) | NC score | 0.554772 (rank : 38) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
UBF1_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 23) | NC score | 0.546838 (rank : 41) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
SOX14_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 24) | NC score | 0.585680 (rank : 16) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O95416, Q3KPH7 | Gene names | SOX14, SOX28 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX14_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 25) | NC score | 0.585745 (rank : 15) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q04892, Q9JLC9, Q9JLD0, Q9QXU4 | Gene names | Sox14, Sox-14 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX17_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 26) | NC score | 0.563966 (rank : 32) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q61473, Q61472, Q62248 | Gene names | Sox17, Sox-17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX7_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 27) | NC score | 0.580998 (rank : 19) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
SOX7_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 28) | NC score | 0.584118 (rank : 17) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P40646, Q9R1T6 | Gene names | Sox7, Sox-7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7 (mSOX7). | |||||
|
SOX18_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 29) | NC score | 0.563082 (rank : 33) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P43680, Q9EQ73 | Gene names | Sox18, Sox-18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
TFAM_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 30) | NC score | 0.610220 (rank : 14) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P40630, P97894, P97906, Q543I8, Q9DBM9 | Gene names | Tfam, Hmgts | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Testis- specific high mobility group protein) (TS-HMG). | |||||
|
SOX17_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 31) | NC score | 0.561309 (rank : 34) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9H6I2 | Gene names | SOX17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX18_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 32) | NC score | 0.567854 (rank : 30) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P35713, Q9NPH8 | Gene names | SOX18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
SOX2_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 33) | NC score | 0.570839 (rank : 24) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P48431, Q14537 | Gene names | SOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX2_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 34) | NC score | 0.570823 (rank : 25) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P48432 | Gene names | Sox2, Sox-2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
CIC_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 35) | NC score | 0.449047 (rank : 70) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CIC_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 36) | NC score | 0.477310 (rank : 66) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
BBX_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 37) | NC score | 0.518870 (rank : 53) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8WY36, Q2TAJ1, Q7L3J8, Q7LBY8, Q8NDB0, Q8WY35, Q9H0J6 | Gene names | BBX, HBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
BBX_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 38) | NC score | 0.528567 (rank : 50) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8VBW5, Q3TZK1, Q6NXY8, Q6PEU3, Q8BQJ7, Q8C7E0, Q8CDQ0, Q8CDV1, Q8VI48, Q8VI49, Q8VI50, Q9CS94 | Gene names | Bbx, Hbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
SOX21_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 39) | NC score | 0.569790 (rank : 26) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9Y651, P35715, Q15504 | Gene names | SOX21, SOX25, SOXA | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-21 (SOX-A). | |||||
|
SOX21_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 40) | NC score | 0.569778 (rank : 27) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q811W0, Q8VH35 | Gene names | Sox21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-21. | |||||
|
TFAM_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 41) | NC score | 0.565197 (rank : 31) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q00059 | Gene names | TFAM, TCF6L2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Mitochondrial transcription factor 1) (MtTF1) (Transcription factor 6-like 2). | |||||
|
SOX15_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 42) | NC score | 0.569388 (rank : 28) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O60248, P35717, Q9Y6W7 | Gene names | SOX15, SOX12, SOX20, SOX26, SOX27 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein (SOX-20 protein) (SOX-12 protein). | |||||
|
SOX1_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 43) | NC score | 0.557172 (rank : 35) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O00570 | Gene names | SOX1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX1_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 44) | NC score | 0.556832 (rank : 36) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P53783 | Gene names | Sox1, Sox-1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX3_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 45) | NC score | 0.553389 (rank : 39) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P41225, P35714, Q9NP49 | Gene names | SOX3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SOX3_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 46) | NC score | 0.556204 (rank : 37) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P53784 | Gene names | Sox3, Sox-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SOX30_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 47) | NC score | 0.511954 (rank : 55) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SOX30_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 48) | NC score | 0.514824 (rank : 54) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SOX8_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 49) | NC score | 0.527075 (rank : 51) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q04886 | Gene names | Sox8, Sox-8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
GCX1_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 50) | NC score | 0.480931 (rank : 65) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96NM4, Q5TE33, Q5TE34, Q5TE35, Q96IC9, Q9BQN5 | Gene names | GCX1, C20orf100 | |||
|
Domain Architecture |

|
|||||
| Description | Granulosa cell HMG box protein 1 (GCX-1). | |||||
|
SOX15_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 51) | NC score | 0.568379 (rank : 29) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P43267, O70204, P70418, Q62246, Q91V00, Q91V43, Q920T1, Q9JLG2 | Gene names | Sox15, Sox-15 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein. | |||||
|
SOX4_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 52) | NC score | 0.533335 (rank : 48) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q06831 | Gene names | Sox4, Sox-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX5_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 53) | NC score | 0.492872 (rank : 60) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX8_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 54) | NC score | 0.527014 (rank : 52) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P57073, Q9NZW2 | Gene names | SOX8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX13_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 55) | NC score | 0.487216 (rank : 62) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 56) | NC score | 0.489889 (rank : 61) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SOX5_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 57) | NC score | 0.493852 (rank : 59) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P35710, O88184, O89018 | Gene names | Sox5, Sox-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
ST18_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 58) | NC score | 0.085170 (rank : 83) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80TY4, Q3UH00, Q3URH9, Q3UVB9, Q3UZN9, Q811B4, Q8K098 | Gene names | St18, Kiaa0535 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18. | |||||
|
SOX12_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 59) | NC score | 0.542176 (rank : 44) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O15370, Q9NUD4 | Gene names | SOX12, SOX22 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-12 protein (SOX-22 protein). | |||||
|
SOX12_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 60) | NC score | 0.542783 (rank : 43) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q04890, P70417, Q6NXL2 | Gene names | Sox12, Sox-12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SOX-12 protein. | |||||
|
SOX9_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 61) | NC score | 0.502430 (rank : 58) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX9_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 62) | NC score | 0.503335 (rank : 57) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q04887, Q8C7L2, Q91ZK2, Q99KQ0 | Gene names | Sox9, Sox-9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
ST18_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 63) | NC score | 0.084145 (rank : 84) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60284 | Gene names | ST18, KIAA0535, ZNF387 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18 (Zinc finger protein 387). | |||||
|
SOX4_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 64) | NC score | 0.533282 (rank : 49) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q06945 | Gene names | SOX4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
CAN3_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 65) | NC score | 0.034143 (rank : 101) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64691, Q9WUC5 | Gene names | Capn3 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3). | |||||
|
SOX11_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 66) | NC score | 0.533973 (rank : 47) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q7M6Y2, O35178, O89036, Q04889, Q80XF0 | Gene names | Sox11, Sox-11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-11. | |||||
|
SRY_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 67) | NC score | 0.549802 (rank : 40) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q05066 | Gene names | SRY, TDF | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
SOX11_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 68) | NC score | 0.534820 (rank : 46) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P35716 | Gene names | SOX11 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX6_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 69) | NC score | 0.482358 (rank : 63) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P35712, Q9BXQ3, Q9BXQ4, Q9BXQ5, Q9H0I8 | Gene names | SOX6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6. | |||||
|
SOX6_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 70) | NC score | 0.481251 (rank : 64) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P40645, Q62250, Q9QWS5 | Gene names | Sox6, Sox-6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6 (SOX-LZ). | |||||
|
SRY_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 71) | NC score | 0.538697 (rank : 45) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q05738 | Gene names | Sry, Tdf, Tdy | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
LEF1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 72) | NC score | 0.360874 (rank : 75) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UJU2, Q9HAZ0 | Gene names | LEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1) (T cell-specific transcription factor 1-alpha) (TCF1-alpha). | |||||
|
LEF1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 73) | NC score | 0.360488 (rank : 76) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P27782 | Gene names | Lef1, Lef-1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1). | |||||
|
REST_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 74) | NC score | 0.045729 (rank : 91) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
HM2L1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 75) | NC score | 0.457380 (rank : 67) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UGU5, O75672, O75673, Q9UMT5 | Gene names | HMG2L1, HMGBCG | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 2-like 1 (Protein HMGBCG). | |||||
|
TCF7_MOUSE
|
||||||
| θ value | 0.125558 (rank : 76) | NC score | 0.355909 (rank : 77) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
NEMO_HUMAN
|
||||||
| θ value | 0.279714 (rank : 77) | NC score | 0.037053 (rank : 95) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y6K9 | Gene names | IKBKG, FIP3, NEMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (FIP-3). | |||||
|
REST_MOUSE
|
||||||
| θ value | 0.279714 (rank : 78) | NC score | 0.047726 (rank : 90) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
TF7L1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 79) | NC score | 0.326422 (rank : 80) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Z1J1, O70450, O70573 | Gene names | Tcf7l1, Tcf3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF-3) (mTCF-3). | |||||
|
CF060_HUMAN
|
||||||
| θ value | 0.47712 (rank : 80) | NC score | 0.043208 (rank : 92) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
HBP1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 81) | NC score | 0.406095 (rank : 71) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O60381, Q8TBM1, Q8TE93, Q96AJ2 | Gene names | HBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
HBP1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 82) | NC score | 0.405626 (rank : 72) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8R316, Q3V0I4, Q8BUS3, Q8C199 | Gene names | Hbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 83) | NC score | 0.036814 (rank : 97) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
TF7L1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 84) | NC score | 0.325734 (rank : 81) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9HCS4, Q53R97, Q6PD70, Q9NP00 | Gene names | TCF7L1, TCF3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF- 3). | |||||
|
TF7L2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 85) | NC score | 0.342534 (rank : 79) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NQB0, O00185, Q9NQB1, Q9NQB2, Q9NQB3, Q9NQB4, Q9NQB5, Q9NQB6, Q9NQB7, Q9ULC2 | Gene names | TCF7L2, TCF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (hTCF-4). | |||||
|
TF7L2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 86) | NC score | 0.343845 (rank : 78) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q924A0, O70574, Q91XP2, Q91XP3, Q91XP4, Q924A1, Q9Z0V3, Q9Z0V4 | Gene names | Tcf7l2, Tcf4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (mTCF-4). | |||||
|
TCF7_HUMAN
|
||||||
| θ value | 0.62314 (rank : 87) | NC score | 0.380469 (rank : 74) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P36402, Q9UKI4 | Gene names | TCF7, TCF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 88) | NC score | 0.031744 (rank : 103) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 0.813845 (rank : 89) | NC score | 0.034711 (rank : 100) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CYLN2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 90) | NC score | 0.036965 (rank : 96) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
CYLN2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 91) | NC score | 0.037571 (rank : 94) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
KIF14_HUMAN
|
||||||
| θ value | 1.38821 (rank : 92) | NC score | 0.017778 (rank : 120) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15058 | Gene names | KIF14, KIAA0042 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF14. | |||||
|
TRI38_HUMAN
|
||||||
| θ value | 1.38821 (rank : 93) | NC score | 0.010012 (rank : 126) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00635 | Gene names | TRIM38, RNF15, RORET | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 38 (RING finger protein 15) (Zinc finger protein RoRet). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 94) | NC score | 0.021326 (rank : 117) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.035160 (rank : 99) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
KIF23_HUMAN
|
||||||
| θ value | 2.36792 (rank : 96) | NC score | 0.019095 (rank : 118) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q02241 | Gene names | KIF23, KNSL5, MKLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF23 (Mitotic kinesin-like protein 1) (Kinesin- like protein 5). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.035703 (rank : 98) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
GOGA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.032925 (rank : 102) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.029614 (rank : 108) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
TRI41_HUMAN
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.011177 (rank : 124) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WV44, Q5BKT0, Q7L484, Q96Q10, Q9BSL8 | Gene names | TRIM41 | |||
|
Domain Architecture |
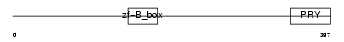
|
|||||
| Description | Tripartite motif-containing protein 41. | |||||
|
UBN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.066358 (rank : 87) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NPG3, Q13079, Q9P1P7 | Gene names | UBN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubinuclein (Ubiquitously expressed nuclear protein) (VT4). | |||||
|
LAMB3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.009194 (rank : 127) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13751, O14947, Q14733, Q9UJK4, Q9UJL1 | Gene names | LAMB3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Laminin B1k chain) (Kalinin B1 chain). | |||||
|
VGFR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | -0.004410 (rank : 129) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17948, O60722, P16057, Q12954 | Gene names | FLT1, FLT, FRT | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor receptor 1 precursor (EC 2.7.10.1) (VEGFR-1) (Vascular permeability factor receptor) (Tyrosine-protein kinase receptor FLT) (Flt-1) (Tyrosine-protein kinase FRT) (Fms-like tyrosine kinase 1). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.027908 (rank : 110) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.022453 (rank : 116) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
K0555_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.037984 (rank : 93) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
NIN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.026908 (rank : 111) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
NOP14_MOUSE
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.022798 (rank : 115) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R3N1 | Gene names | Nop14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
CLC4F_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.018287 (rank : 119) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 572 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70194, Q8BLZ8 | Gene names | Clec4f, Clecsf13, Kclr | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member F (C-type lectin superfamily member 13) (C-type lectin 13) (Kupffer cell receptor). | |||||
|
DIAP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.013908 (rank : 121) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.023665 (rank : 113) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
PEPL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.029846 (rank : 107) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
RC3H1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.030057 (rank : 106) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q4VGL6, Q69Z31 | Gene names | Rc3h1, Gm551, Kiaa2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1) (Sanroque protein). | |||||
|
TAOK3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | -0.000492 (rank : 128) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1458 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BYC6, Q3V3B3 | Gene names | Taok3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO3 (EC 2.7.11.1) (Thousand and one amino acid protein 3). | |||||
|
TEX9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.022981 (rank : 114) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N6V9 | Gene names | TEX9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein. | |||||
|
TRI32_MOUSE
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.010769 (rank : 125) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CH72, Q8K055 | Gene names | Trim32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-). | |||||
|
U2AFM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.025566 (rank : 112) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
GOPC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.013226 (rank : 122) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HD26, Q59FS4, Q969U8 | Gene names | GOPC, CAL, FIG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi-associated PDZ and coiled-coil motif-containing protein (PDZ protein interacting specifically with TC10) (PIST) (CFTR-associated ligand) (Fused in glioblastoma). | |||||
|
GOPC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.012920 (rank : 123) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BH60, Q8BSV4, Q920R1, Q9ET11 | Gene names | Gopc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi-associated PDZ and coiled-coil motif-containing protein (PDZ protein interacting specifically with TC10) (PIST). | |||||
|
LIPA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.030797 (rank : 105) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.031510 (rank : 104) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
LIPA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.029016 (rank : 109) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
MYT1L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.051930 (rank : 88) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
MYT1L_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.050985 (rank : 89) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
MYT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.073947 (rank : 85) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
MYT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.071079 (rank : 86) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
PMS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.265386 (rank : 82) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
SOX10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.450976 (rank : 69) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
SOX10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.450987 (rank : 68) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q04888, O08518, O09141, O54856, P70416 | Gene names | Sox10, Sox-10, Sox21 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10 (SOX-21) (Transcription factor SOX-M). | |||||
|
HM20B_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.75757e-126 (rank : 1) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | Q9Z104, Q9NSF7, Q9NSF8 | Gene names | Hmg20b, Braf35, Hmgx2, Smarce1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35). | |||||
|
HM20B_HUMAN
|
||||||
| NC score | 0.994391 (rank : 2) | θ value | 1.17455e-116 (rank : 2) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q9P0W2, Q6IBP8, Q8NBD5, Q9HD21, Q9Y491, Q9Y4A2 | Gene names | HMG20B, BRAF35, HMGX2, SMARCE1R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35) (Sox-like transcriptional factor). | |||||
|
HM20A_HUMAN
|
||||||
| NC score | 0.889140 (rank : 3) | θ value | 2.09745e-73 (rank : 3) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9NP66, Q53G31, Q9NSF6 | Gene names | HMG20A, HMGX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (HMG domain-containing protein HMGX1). | |||||
|
HM20A_MOUSE
|
||||||
| NC score | 0.886388 (rank : 4) | θ value | 7.97034e-73 (rank : 4) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9DC33, Q3LSF9, Q6NV87, Q8BSK1, Q8C3C1, Q8CAA0, Q9CYG2 | Gene names | Hmg20a, Ibraf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (iBRAF) (Inhibitor of BRAF35) (HMG domain protein HMGX1). | |||||
|
HMGB3_MOUSE
|
||||||
| NC score | 0.708126 (rank : 5) | θ value | 7.34386e-10 (rank : 6) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O54879 | Gene names | Hmgb3, Hmg2a, Hmg4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
HMGB3_HUMAN
|
||||||
| NC score | 0.707777 (rank : 6) | θ value | 4.30538e-10 (rank : 5) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O15347, O95556, Q6NS40 | Gene names | HMGB3, HMG2A, HMG4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
HMGB2_HUMAN
|
||||||
| NC score | 0.695659 (rank : 7) | θ value | 4.76016e-09 (rank : 12) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P26583 | Gene names | HMGB2, HMG2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
HMGB2_MOUSE
|
||||||
| NC score | 0.690357 (rank : 8) | θ value | 8.11959e-09 (rank : 17) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P30681, Q3UXT1, Q9EQD5 | Gene names | Hmgb2, Hmg2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
HMGB1_MOUSE
|
||||||
| NC score | 0.686531 (rank : 9) | θ value | 1.25267e-09 (rank : 8) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P63158, P07155, P27109, P27428 | Gene names | Hmgb1, Hmg-1, Hmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HMGB1_HUMAN
|
||||||
| NC score | 0.685974 (rank : 10) | θ value | 1.25267e-09 (rank : 7) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P09429, Q6IBE1 | Gene names | HMGB1, HMG1 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HMG1X_HUMAN
|
||||||
| NC score | 0.657647 (rank : 11) | θ value | 5.26297e-08 (rank : 18) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UGV6 | Gene names | HMG1L10 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 1-like 10 (HMG-1L10). | |||||
|
SSRP1_MOUSE
|
||||||
| NC score | 0.642269 (rank : 12) | θ value | 1.17247e-07 (rank : 20) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
SSRP1_HUMAN
|
||||||
| NC score | 0.641945 (rank : 13) | θ value | 1.53129e-07 (rank : 21) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
TFAM_MOUSE
|
||||||
| NC score | 0.610220 (rank : 14) | θ value | 1.43324e-05 (rank : 30) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P40630, P97894, P97906, Q543I8, Q9DBM9 | Gene names | Tfam, Hmgts | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Testis- specific high mobility group protein) (TS-HMG). | |||||
|
SOX14_MOUSE
|
||||||
| NC score | 0.585745 (rank : 15) | θ value | 1.29631e-06 (rank : 25) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q04892, Q9JLC9, Q9JLD0, Q9QXU4 | Gene names | Sox14, Sox-14 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX14_HUMAN
|
||||||
| NC score | 0.585680 (rank : 16) | θ value | 1.29631e-06 (rank : 24) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O95416, Q3KPH7 | Gene names | SOX14, SOX28 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX7_MOUSE
|
||||||
| NC score | 0.584118 (rank : 17) | θ value | 8.40245e-06 (rank : 28) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P40646, Q9R1T6 | Gene names | Sox7, Sox-7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7 (mSOX7). | |||||
|
CN092_HUMAN
|
||||||
| NC score | 0.581294 (rank : 18) | θ value | 6.21693e-09 (rank : 15) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O94842 | Gene names | C14orf92, KIAA0737 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
SOX7_HUMAN
|
||||||
| NC score | 0.580998 (rank : 19) | θ value | 6.43352e-06 (rank : 27) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
SMCE1_HUMAN
|
||||||
| NC score | 0.578669 (rank : 20) | θ value | 1.63604e-09 (rank : 9) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
TOX_MOUSE
|
||||||
| NC score | 0.575327 (rank : 21) | θ value | 4.76016e-09 (rank : 14) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q66JW3, Q8BKH9, Q8BYQ5, Q8R4H0 | Gene names | Tox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
TOX_HUMAN
|
||||||
| NC score | 0.572948 (rank : 22) | θ value | 6.21693e-09 (rank : 16) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O94900, Q96AV5 | Gene names | TOX, KIAA0808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
CN092_MOUSE
|
||||||
| NC score | 0.571015 (rank : 23) | θ value | 2.13673e-09 (rank : 11) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
SOX2_HUMAN
|
||||||
| NC score | 0.570839 (rank : 24) | θ value | 4.1701e-05 (rank : 33) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P48431, Q14537 | Gene names | SOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX2_MOUSE
|
||||||
| NC score | 0.570823 (rank : 25) | θ value | 4.1701e-05 (rank : 34) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P48432 | Gene names | Sox2, Sox-2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX21_HUMAN
|
||||||
| NC score | 0.569790 (rank : 26) | θ value | 7.1131e-05 (rank : 39) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9Y651, P35715, Q15504 | Gene names | SOX21, SOX25, SOXA | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-21 (SOX-A). | |||||
|
SOX21_MOUSE
|
||||||
| NC score | 0.569778 (rank : 27) | θ value | 7.1131e-05 (rank : 40) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q811W0, Q8VH35 | Gene names | Sox21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-21. | |||||
|
SOX15_HUMAN
|
||||||
| NC score | 0.569388 (rank : 28) | θ value | 9.29e-05 (rank : 42) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O60248, P35717, Q9Y6W7 | Gene names | SOX15, SOX12, SOX20, SOX26, SOX27 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein (SOX-20 protein) (SOX-12 protein). | |||||
|
SOX15_MOUSE
|
||||||
| NC score | 0.568379 (rank : 29) | θ value | 0.000602161 (rank : 51) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P43267, O70204, P70418, Q62246, Q91V00, Q91V43, Q920T1, Q9JLG2 | Gene names | Sox15, Sox-15 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein. | |||||
|
SOX18_HUMAN
|
||||||
| NC score | 0.567854 (rank : 30) | θ value | 2.44474e-05 (rank : 32) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P35713, Q9NPH8 | Gene names | SOX18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
TFAM_HUMAN
|
||||||
| NC score | 0.565197 (rank : 31) | θ value | 7.1131e-05 (rank : 41) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q00059 | Gene names | TFAM, TCF6L2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Mitochondrial transcription factor 1) (MtTF1) (Transcription factor 6-like 2). | |||||
|
SOX17_MOUSE
|
||||||
| NC score | 0.563966 (rank : 32) | θ value | 2.21117e-06 (rank : 26) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q61473, Q61472, Q62248 | Gene names | Sox17, Sox-17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX18_MOUSE
|
||||||
| NC score | 0.563082 (rank : 33) | θ value | 1.43324e-05 (rank : 29) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P43680, Q9EQ73 | Gene names | Sox18, Sox-18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
SOX17_HUMAN
|
||||||
| NC score | 0.561309 (rank : 34) | θ value | 2.44474e-05 (rank : 31) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9H6I2 | Gene names | SOX17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX1_HUMAN
|
||||||
| NC score | 0.557172 (rank : 35) | θ value | 9.29e-05 (rank : 43) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O00570 | Gene names | SOX1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX1_MOUSE
|
||||||
| NC score | 0.556832 (rank : 36) | θ value | 9.29e-05 (rank : 44) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P53783 | Gene names | Sox1, Sox-1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX3_MOUSE
|
||||||
| NC score | 0.556204 (rank : 37) | θ value | 9.29e-05 (rank : 46) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P53784 | Gene names | Sox3, Sox-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.554772 (rank : 38) | θ value | 2.61198e-07 (rank : 22) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
SOX3_HUMAN
|
||||||
| NC score | 0.553389 (rank : 39) | θ value | 9.29e-05 (rank : 45) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P41225, P35714, Q9NP49 | Gene names | SOX3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SRY_HUMAN
|
||||||
| NC score | 0.549802 (rank : 40) | θ value | 0.00509761 (rank : 67) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q05066 | Gene names | SRY, TDF | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
UBF1_MOUSE
|
||||||
| NC score | 0.546838 (rank : 41) | θ value | 2.61198e-07 (rank : 23) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
SMCE1_MOUSE
|
||||||
| NC score | 0.546197 (rank : 42) | θ value | 1.63604e-09 (rank : 10) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
SOX12_MOUSE
|
||||||
| NC score | 0.542783 (rank : 43) | θ value | 0.00228821 (rank : 60) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q04890, P70417, Q6NXL2 | Gene names | Sox12, Sox-12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SOX-12 protein. | |||||
|
SOX12_HUMAN
|
||||||
| NC score | 0.542176 (rank : 44) | θ value | 0.00228821 (rank : 59) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O15370, Q9NUD4 | Gene names | SOX12, SOX22 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-12 protein (SOX-22 protein). | |||||
|
SRY_MOUSE
|
||||||
| NC score | 0.538697 (rank : 45) | θ value | 0.00869519 (rank : 71) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q05738 | Gene names | Sry, Tdf, Tdy | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
SOX11_HUMAN
|
||||||
| NC score | 0.534820 (rank : 46) | θ value | 0.00665767 (rank : 68) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P35716 | Gene names | SOX11 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX11_MOUSE
|
||||||
| NC score | 0.533973 (rank : 47) | θ value | 0.00509761 (rank : 66) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q7M6Y2, O35178, O89036, Q04889, Q80XF0 | Gene names | Sox11, Sox-11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX4_MOUSE
|
||||||
| NC score | 0.533335 (rank : 48) | θ value | 0.00102713 (rank : 52) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q06831 | Gene names | Sox4, Sox-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX4_HUMAN
|
||||||
| NC score | 0.533282 (rank : 49) | θ value | 0.00390308 (rank : 64) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q06945 | Gene names | SOX4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
BBX_MOUSE
|
||||||
| NC score | 0.528567 (rank : 50) | θ value | 7.1131e-05 (rank : 38) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8VBW5, Q3TZK1, Q6NXY8, Q6PEU3, Q8BQJ7, Q8C7E0, Q8CDQ0, Q8CDV1, Q8VI48, Q8VI49, Q8VI50, Q9CS94 | Gene names | Bbx, Hbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
SOX8_MOUSE
|
||||||
| NC score | 0.527075 (rank : 51) | θ value | 0.000270298 (rank : 49) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q04886 | Gene names | Sox8, Sox-8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX8_HUMAN
|
||||||
| NC score | 0.527014 (rank : 52) | θ value | 0.00102713 (rank : 54) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P57073, Q9NZW2 | Gene names | SOX8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
BBX_HUMAN
|
||||||
| NC score | 0.518870 (rank : 53) | θ value | 7.1131e-05 (rank : 37) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8WY36, Q2TAJ1, Q7L3J8, Q7LBY8, Q8NDB0, Q8WY35, Q9H0J6 | Gene names | BBX, HBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
SOX30_MOUSE
|
||||||
| NC score | 0.514824 (rank : 54) | θ value | 0.000121331 (rank : 48) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SOX30_HUMAN
|
||||||
| NC score | 0.511954 (rank : 55) | θ value | 0.000121331 (rank : 47) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SP100_HUMAN
|
||||||
| NC score | 0.510639 (rank : 56) | θ value | 4.76016e-09 (rank : 13) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P23497, O75450, Q13343, Q96F70, Q96T24, Q96T95, Q9NP33, Q9UE32 | Gene names | SP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein) (Lysp100b). | |||||
|
SOX9_MOUSE
|
||||||
| NC score | 0.503335 (rank : 57) | θ value | 0.00228821 (rank : 62) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q04887, Q8C7L2, Q91ZK2, Q99KQ0 | Gene names | Sox9, Sox-9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX9_HUMAN
|
||||||
| NC score | 0.502430 (rank : 58) | θ value | 0.00228821 (rank : 61) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX5_MOUSE
|
||||||
| NC score | 0.493852 (rank : 59) | θ value | 0.00175202 (rank : 57) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P35710, O88184, O89018 | Gene names | Sox5, Sox-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX5_HUMAN
|
||||||
| NC score | 0.492872 (rank : 60) | θ value | 0.00102713 (rank : 53) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.489889 (rank : 61) | θ value | 0.00134147 (rank : 56) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SOX13_HUMAN
|
||||||
| NC score | 0.487216 (rank : 62) | θ value | 0.00134147 (rank : 55) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
SOX6_HUMAN
|
||||||
| NC score | 0.482358 (rank : 63) | θ value | 0.00869519 (rank : 69) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P35712, Q9BXQ3, Q9BXQ4, Q9BXQ5, Q9H0I8 | Gene names | SOX6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6. | |||||
|
SOX6_MOUSE
|
||||||
| NC score | 0.481251 (rank : 64) | θ value | 0.00869519 (rank : 70) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P40645, Q62250, Q9QWS5 | Gene names | Sox6, Sox-6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6 (SOX-LZ). | |||||
|
GCX1_HUMAN
|
||||||
| NC score | 0.480931 (rank : 65) | θ value | 0.000461057 (rank : 50) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96NM4, Q5TE33, Q5TE34, Q5TE35, Q96IC9, Q9BQN5 | Gene names | GCX1, C20orf100 | |||
|
Domain Architecture |

|
|||||
| Description | Granulosa cell HMG box protein 1 (GCX-1). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.477310 (rank : 66) | θ value | 5.44631e-05 (rank : 36) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
HM2L1_HUMAN
|
||||||
| NC score | 0.457380 (rank : 67) | θ value | 0.0961366 (rank : 75) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UGU5, O75672, O75673, Q9UMT5 | Gene names | HMG2L1, HMGBCG | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 2-like 1 (Protein HMGBCG). | |||||
|
SOX10_MOUSE
|
||||||
| NC score | 0.450987 (rank : 68) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q04888, O08518, O09141, O54856, P70416 | Gene names | Sox10, Sox-10, Sox21 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10 (SOX-21) (Transcription factor SOX-M). | |||||
|
SOX10_HUMAN
|
||||||
| NC score | 0.450976 (rank : 69) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.449047 (rank : 70) | θ value | 5.44631e-05 (rank : 35) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
HBP1_HUMAN
|
||||||
| NC score | 0.406095 (rank : 71) | θ value | 0.47712 (rank : 81) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O60381, Q8TBM1, Q8TE93, Q96AJ2 | Gene names | HBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
HBP1_MOUSE
|
||||||
| NC score | 0.405626 (rank : 72) | θ value | 0.47712 (rank : 82) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8R316, Q3V0I4, Q8BUS3, Q8C199 | Gene names | Hbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
PB1_HUMAN
|
||||||
| NC score | 0.397584 (rank : 73) | θ value | 5.26297e-08 (rank : 19) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
TCF7_HUMAN
|
||||||
| NC score | 0.380469 (rank : 74) | θ value | 0.62314 (rank : 87) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P36402, Q9UKI4 | Gene names | TCF7, TCF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
LEF1_HUMAN
|
||||||
| NC score | 0.360874 (rank : 75) | θ value | 0.0431538 (rank : 72) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UJU2, Q9HAZ0 | Gene names | LEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1) (T cell-specific transcription factor 1-alpha) (TCF1-alpha). | |||||
|
LEF1_MOUSE
|
||||||
| NC score | 0.360488 (rank : 76) | θ value | 0.0431538 (rank : 73) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P27782 | Gene names | Lef1, Lef-1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1). | |||||
|
TCF7_MOUSE
|
||||||
| NC score | 0.355909 (rank : 77) | θ value | 0.125558 (rank : 76) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
TF7L2_MOUSE
|
||||||
| NC score | 0.343845 (rank : 78) | θ value | 0.47712 (rank : 86) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q924A0, O70574, Q91XP2, Q91XP3, Q91XP4, Q924A1, Q9Z0V3, Q9Z0V4 | Gene names | Tcf7l2, Tcf4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (mTCF-4). | |||||
|
TF7L2_HUMAN
|
||||||
| NC score | 0.342534 (rank : 79) | θ value | 0.47712 (rank : 85) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NQB0, O00185, Q9NQB1, Q9NQB2, Q9NQB3, Q9NQB4, Q9NQB5, Q9NQB6, Q9NQB7, Q9ULC2 | Gene names | TCF7L2, TCF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (hTCF-4). | |||||
|
TF7L1_MOUSE
|
||||||
| NC score | 0.326422 (rank : 80) | θ value | 0.365318 (rank : 79) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Z1J1, O70450, O70573 | Gene names | Tcf7l1, Tcf3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF-3) (mTCF-3). | |||||
|
TF7L1_HUMAN
|
||||||
| NC score | 0.325734 (rank : 81) | θ value | 0.47712 (rank : 84) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9HCS4, Q53R97, Q6PD70, Q9NP00 | Gene names | TCF7L1, TCF3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF- 3). | |||||
|
PMS1_HUMAN
|
||||||
| NC score | 0.265386 (rank : 82) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
ST18_MOUSE
|
||||||
| NC score | 0.085170 (rank : 83) | θ value | 0.00175202 (rank : 58) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80TY4, Q3UH00, Q3URH9, Q3UVB9, Q3UZN9, Q811B4, Q8K098 | Gene names | St18, Kiaa0535 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18. | |||||
|
ST18_HUMAN
|
||||||
| NC score | 0.084145 (rank : 84) | θ value | 0.00228821 (rank : 63) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60284 | Gene names | ST18, KIAA0535, ZNF387 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18 (Zinc finger protein 387). | |||||
|
MYT1_HUMAN
|
||||||
| NC score | 0.073947 (rank : 85) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.071079 (rank : 86) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
UBN1_HUMAN
|
||||||
| NC score | 0.066358 (rank : 87) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NPG3, Q13079, Q9P1P7 | Gene names | UBN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubinuclein (Ubiquitously expressed nuclear protein) (VT4). | |||||
|
MYT1L_HUMAN
|
||||||
| NC score | 0.051930 (rank : 88) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
MYT1L_MOUSE
|
||||||
| NC score | 0.050985 (rank : 89) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.047726 (rank : 90) | θ value | 0.279714 (rank : 78) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.045729 (rank : 91) | θ value | 0.0563607 (rank : 74) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.043208 (rank : 92) | θ value | 0.47712 (rank : 80) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
K0555_HUMAN
|
||||||
| NC score | 0.037984 (rank : 93) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
CYLN2_MOUSE
|
||||||
| NC score | 0.037571 (rank : 94) | θ value | 1.38821 (rank : 91) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
NEMO_HUMAN
|
||||||
| NC score | 0.037053 (rank : 95) | θ value | 0.279714 (rank : 77) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y6K9 | Gene names | IKBKG, FIP3, NEMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (FIP-3). | |||||
|
CYLN2_HUMAN
|
||||||
| NC score | 0.036965 (rank : 96) | θ value | 1.06291 (rank : 90) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.036814 (rank : 97) | θ value | 0.47712 (rank : 83) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.035703 (rank : 98) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.035160 (rank : 99) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.034711 (rank : 100) | θ value | 0.813845 (rank : 89) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CAN3_MOUSE
|
||||||
| NC score | 0.034143 (rank : 101) | θ value | 0.00509761 (rank : 65) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64691, Q9WUC5 | Gene names | Capn3 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3). | |||||
|
GOGA1_MOUSE
|
||||||
| NC score | 0.032925 (rank : 102) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.031744 (rank : 103) | θ value | 0.813845 (rank : 88) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.031510 (rank : 104) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
LIPA2_HUMAN
|
||||||
| NC score | 0.030797 (rank : 105) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
RC3H1_MOUSE
|
||||||
| NC score | 0.030057 (rank : 106) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q4VGL6, Q69Z31 | Gene names | Rc3h1, Gm551, Kiaa2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1) (Sanroque protein). | |||||
|
PEPL_MOUSE
|
||||||
| NC score | 0.029846 (rank : 107) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.029614 (rank : 108) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
LIPA3_HUMAN
|
||||||
| NC score | 0.029016 (rank : 109) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.027908 (rank : 110) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.026908 (rank : 111) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
U2AFM_HUMAN
|
||||||
| NC score | 0.025566 (rank : 112) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.023665 (rank : 113) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
TEX9_HUMAN
|
||||||
| NC score | 0.022981 (rank : 114) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N6V9 | Gene names | TEX9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein. | |||||
|
NOP14_MOUSE
|
||||||
| NC score | 0.022798 (rank : 115) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R3N1 | Gene names | Nop14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.022453 (rank : 116) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.021326 (rank : 117) | θ value | 1.81305 (rank : 94) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
KIF23_HUMAN
|
||||||
| NC score | 0.019095 (rank : 118) | θ value | 2.36792 (rank : 96) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q02241 | Gene names | KIF23, KNSL5, MKLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF23 (Mitotic kinesin-like protein 1) (Kinesin- like protein 5). | |||||
|
CLC4F_MOUSE
|
||||||
| NC score | 0.018287 (rank : 119) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 572 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70194, Q8BLZ8 | Gene names | Clec4f, Clecsf13, Kclr | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member F (C-type lectin superfamily member 13) (C-type lectin 13) (Kupffer cell receptor). | |||||
|
KIF14_HUMAN
|
||||||
| NC score | 0.017778 (rank : 120) | θ value | 1.38821 (rank : 92) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15058 | Gene names | KIF14, KIAA0042 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF14. | |||||
|
DIAP2_HUMAN
|
||||||
| NC score | 0.013908 (rank : 121) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
GOPC_HUMAN
|
||||||
| NC score | 0.013226 (rank : 122) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HD26, Q59FS4, Q969U8 | Gene names | GOPC, CAL, FIG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi-associated PDZ and coiled-coil motif-containing protein (PDZ protein interacting specifically with TC10) (PIST) (CFTR-associated ligand) (Fused in glioblastoma). | |||||
|
GOPC_MOUSE
|
||||||
| NC score | 0.012920 (rank : 123) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BH60, Q8BSV4, Q920R1, Q9ET11 | Gene names | Gopc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi-associated PDZ and coiled-coil motif-containing protein (PDZ protein interacting specifically with TC10) (PIST). | |||||
|
TRI41_HUMAN
|
||||||
| NC score | 0.011177 (rank : 124) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WV44, Q5BKT0, Q7L484, Q96Q10, Q9BSL8 | Gene names | TRIM41 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 41. | |||||
|
TRI32_MOUSE
|
||||||
| NC score | 0.010769 (rank : 125) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CH72, Q8K055 | Gene names | Trim32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-). | |||||
|
TRI38_HUMAN
|
||||||
| NC score | 0.010012 (rank : 126) | θ value | 1.38821 (rank : 93) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00635 | Gene names | TRIM38, RNF15, RORET | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 38 (RING finger protein 15) (Zinc finger protein RoRet). | |||||
|
LAMB3_HUMAN
|
||||||
| NC score | 0.009194 (rank : 127) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13751, O14947, Q14733, Q9UJK4, Q9UJL1 | Gene names | LAMB3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Laminin B1k chain) (Kalinin B1 chain). | |||||
|
TAOK3_MOUSE
|
||||||
| NC score | -0.000492 (rank : 128) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1458 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BYC6, Q3V3B3 | Gene names | Taok3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO3 (EC 2.7.11.1) (Thousand and one amino acid protein 3). | |||||
|
VGFR1_HUMAN
|
||||||
| NC score | -0.004410 (rank : 129) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 122 | Target Neighborhood Hits | 1109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17948, O60722, P16057, Q12954 | Gene names | FLT1, FLT, FRT | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor receptor 1 precursor (EC 2.7.10.1) (VEGFR-1) (Vascular permeability factor receptor) (Tyrosine-protein kinase receptor FLT) (Flt-1) (Tyrosine-protein kinase FRT) (Fms-like tyrosine kinase 1). | |||||