Please be patient as the page loads
|
LEF1_HUMAN
|
||||||
| SwissProt Accessions | Q9UJU2, Q9HAZ0 | Gene names | LEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1) (T cell-specific transcription factor 1-alpha) (TCF1-alpha). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LEF1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 150 | |
| SwissProt Accessions | Q9UJU2, Q9HAZ0 | Gene names | LEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1) (T cell-specific transcription factor 1-alpha) (TCF1-alpha). | |||||
|
LEF1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.997231 (rank : 2) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 141 | |
| SwissProt Accessions | P27782 | Gene names | Lef1, Lef-1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1). | |||||
|
TF7L2_MOUSE
|
||||||
| θ value | 4.32305e-111 (rank : 3) | NC score | 0.971259 (rank : 3) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q924A0, O70574, Q91XP2, Q91XP3, Q91XP4, Q924A1, Q9Z0V3, Q9Z0V4 | Gene names | Tcf7l2, Tcf4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (mTCF-4). | |||||
|
TF7L2_HUMAN
|
||||||
| θ value | 7.14227e-106 (rank : 4) | NC score | 0.963767 (rank : 4) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9NQB0, O00185, Q9NQB1, Q9NQB2, Q9NQB3, Q9NQB4, Q9NQB5, Q9NQB6, Q9NQB7, Q9ULC2 | Gene names | TCF7L2, TCF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (hTCF-4). | |||||
|
TF7L1_HUMAN
|
||||||
| θ value | 2.01746e-92 (rank : 5) | NC score | 0.957551 (rank : 5) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9HCS4, Q53R97, Q6PD70, Q9NP00 | Gene names | TCF7L1, TCF3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF- 3). | |||||
|
TF7L1_MOUSE
|
||||||
| θ value | 4.96919e-91 (rank : 6) | NC score | 0.956415 (rank : 6) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Z1J1, O70450, O70573 | Gene names | Tcf7l1, Tcf3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF-3) (mTCF-3). | |||||
|
TCF7_MOUSE
|
||||||
| θ value | 9.41505e-74 (rank : 7) | NC score | 0.930985 (rank : 8) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
TCF7_HUMAN
|
||||||
| θ value | 6.32992e-62 (rank : 8) | NC score | 0.937600 (rank : 7) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P36402, Q9UKI4 | Gene names | TCF7, TCF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 9) | NC score | 0.527309 (rank : 50) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CIC_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 10) | NC score | 0.554293 (rank : 49) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
SOX6_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 11) | NC score | 0.584684 (rank : 25) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P35712, Q9BXQ3, Q9BXQ4, Q9BXQ5, Q9H0I8 | Gene names | SOX6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6. | |||||
|
SOX6_MOUSE
|
||||||
| θ value | 1.63604e-09 (rank : 12) | NC score | 0.584027 (rank : 26) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P40645, Q62250, Q9QWS5 | Gene names | Sox6, Sox-6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6 (SOX-LZ). | |||||
|
SOX5_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 13) | NC score | 0.579940 (rank : 29) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX5_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 14) | NC score | 0.581667 (rank : 27) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P35710, O88184, O89018 | Gene names | Sox5, Sox-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX30_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 15) | NC score | 0.605070 (rank : 11) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SOX30_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 16) | NC score | 0.606068 (rank : 9) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SOX13_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 17) | NC score | 0.575117 (rank : 36) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 18) | NC score | 0.571257 (rank : 37) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SOX7_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 19) | NC score | 0.593599 (rank : 15) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
SOX7_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 20) | NC score | 0.597534 (rank : 14) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P40646, Q9R1T6 | Gene names | Sox7, Sox-7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7 (mSOX7). | |||||
|
SRY_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 21) | NC score | 0.593058 (rank : 16) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q05738 | Gene names | Sry, Tdf, Tdy | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
SOX2_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 22) | NC score | 0.586402 (rank : 20) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P48431, Q14537 | Gene names | SOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX2_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 23) | NC score | 0.586050 (rank : 21) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P48432 | Gene names | Sox2, Sox-2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX8_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 24) | NC score | 0.581349 (rank : 28) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P57073, Q9NZW2 | Gene names | SOX8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX8_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 25) | NC score | 0.578047 (rank : 31) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q04886 | Gene names | Sox8, Sox-8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX12_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 26) | NC score | 0.602293 (rank : 13) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O15370, Q9NUD4 | Gene names | SOX12, SOX22 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-12 protein (SOX-22 protein). | |||||
|
SOX12_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 27) | NC score | 0.603747 (rank : 12) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q04890, P70417, Q6NXL2 | Gene names | Sox12, Sox-12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SOX-12 protein. | |||||
|
SOX14_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 28) | NC score | 0.587959 (rank : 18) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O95416, Q3KPH7 | Gene names | SOX14, SOX28 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX14_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 29) | NC score | 0.588035 (rank : 17) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q04892, Q9JLC9, Q9JLD0, Q9QXU4 | Gene names | Sox14, Sox-14 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX11_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 30) | NC score | 0.587319 (rank : 19) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P35716 | Gene names | SOX11 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX11_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 31) | NC score | 0.585804 (rank : 22) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q7M6Y2, O35178, O89036, Q04889, Q80XF0 | Gene names | Sox11, Sox-11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX21_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 32) | NC score | 0.585728 (rank : 23) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Y651, P35715, Q15504 | Gene names | SOX21, SOX25, SOXA | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-21 (SOX-A). | |||||
|
SOX21_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 33) | NC score | 0.585699 (rank : 24) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q811W0, Q8VH35 | Gene names | Sox21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-21. | |||||
|
SRY_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 34) | NC score | 0.605206 (rank : 10) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q05066 | Gene names | SRY, TDF | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
SOX17_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 35) | NC score | 0.575720 (rank : 34) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9H6I2 | Gene names | SOX17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX17_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 36) | NC score | 0.575438 (rank : 35) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q61473, Q61472, Q62248 | Gene names | Sox17, Sox-17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX4_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 37) | NC score | 0.579049 (rank : 30) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q06831 | Gene names | Sox4, Sox-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX10_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 38) | NC score | 0.559922 (rank : 45) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
SOX9_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 39) | NC score | 0.560619 (rank : 44) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX9_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 40) | NC score | 0.561262 (rank : 43) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q04887, Q8C7L2, Q91ZK2, Q99KQ0 | Gene names | Sox9, Sox-9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX10_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 41) | NC score | 0.557793 (rank : 47) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q04888, O08518, O09141, O54856, P70416 | Gene names | Sox10, Sox-10, Sox21 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10 (SOX-21) (Transcription factor SOX-M). | |||||
|
SOX1_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 42) | NC score | 0.569653 (rank : 38) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O00570 | Gene names | SOX1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX1_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 43) | NC score | 0.569105 (rank : 40) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P53783 | Gene names | Sox1, Sox-1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX4_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 44) | NC score | 0.577903 (rank : 32) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q06945 | Gene names | SOX4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX18_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 45) | NC score | 0.576156 (rank : 33) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P43680, Q9EQ73 | Gene names | Sox18, Sox-18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
SOX15_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 46) | NC score | 0.566198 (rank : 41) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P43267, O70204, P70418, Q62246, Q91V00, Q91V43, Q920T1, Q9JLG2 | Gene names | Sox15, Sox-15 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein. | |||||
|
SOX3_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 47) | NC score | 0.555964 (rank : 48) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P41225, P35714, Q9NP49 | Gene names | SOX3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SOX3_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 48) | NC score | 0.559002 (rank : 46) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P53784 | Gene names | Sox3, Sox-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SOX15_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 49) | NC score | 0.562293 (rank : 42) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | O60248, P35717, Q9Y6W7 | Gene names | SOX15, SOX12, SOX20, SOX26, SOX27 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein (SOX-20 protein) (SOX-12 protein). | |||||
|
SOX18_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 50) | NC score | 0.569386 (rank : 39) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P35713, Q9NPH8 | Gene names | SOX18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
BBX_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 51) | NC score | 0.484139 (rank : 54) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8WY36, Q2TAJ1, Q7L3J8, Q7LBY8, Q8NDB0, Q8WY35, Q9H0J6 | Gene names | BBX, HBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
BBX_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 52) | NC score | 0.487445 (rank : 53) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8VBW5, Q3TZK1, Q6NXY8, Q6PEU3, Q8BQJ7, Q8C7E0, Q8CDQ0, Q8CDV1, Q8VI48, Q8VI49, Q8VI50, Q9CS94 | Gene names | Bbx, Hbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
HBP1_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 53) | NC score | 0.507406 (rank : 52) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O60381, Q8TBM1, Q8TE93, Q96AJ2 | Gene names | HBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
HBP1_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 54) | NC score | 0.508913 (rank : 51) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8R316, Q3V0I4, Q8BUS3, Q8C199 | Gene names | Hbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 55) | NC score | 0.069688 (rank : 87) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
ANX11_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 56) | NC score | 0.034493 (rank : 112) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P50995 | Gene names | ANXA11, ANX11 | |||
|
Domain Architecture |
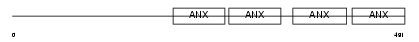
|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50) (56 kDa autoantigen). | |||||
|
HM20A_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 57) | NC score | 0.356240 (rank : 57) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9NP66, Q53G31, Q9NSF6 | Gene names | HMG20A, HMGX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (HMG domain-containing protein HMGX1). | |||||
|
HM20A_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 58) | NC score | 0.355349 (rank : 58) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9DC33, Q3LSF9, Q6NV87, Q8BSK1, Q8C3C1, Q8CAA0, Q9CYG2 | Gene names | Hmg20a, Ibraf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (iBRAF) (Inhibitor of BRAF35) (HMG domain protein HMGX1). | |||||
|
MEF2B_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 59) | NC score | 0.066142 (rank : 90) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 60) | NC score | 0.111594 (rank : 83) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
HXB3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 61) | NC score | 0.031731 (rank : 113) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P14651, O95615, P17484 | Gene names | HOXB3, HOX2G | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2G) (Hox-2.7). | |||||
|
HM20B_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 62) | NC score | 0.364272 (rank : 55) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9P0W2, Q6IBP8, Q8NBD5, Q9HD21, Q9Y491, Q9Y4A2 | Gene names | HMG20B, BRAF35, HMGX2, SMARCE1R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35) (Sox-like transcriptional factor). | |||||
|
HM20B_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 63) | NC score | 0.360874 (rank : 56) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Z104, Q9NSF7, Q9NSF8 | Gene names | Hmg20b, Braf35, Hmgx2, Smarce1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35). | |||||
|
GCX1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 64) | NC score | 0.151385 (rank : 78) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96NM4, Q5TE33, Q5TE34, Q5TE35, Q96IC9, Q9BQN5 | Gene names | GCX1, C20orf100 | |||
|
Domain Architecture |

|
|||||
| Description | Granulosa cell HMG box protein 1 (GCX-1). | |||||
|
HXB3_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 65) | NC score | 0.027214 (rank : 118) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P09026, P10285, Q4PJ06, Q61680 | Gene names | Hoxb3, Hox-2.7, Hoxb-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2.7) (Homeobox protein MH-23). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 66) | NC score | 0.095937 (rank : 85) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SSRP1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 67) | NC score | 0.339519 (rank : 59) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
CN092_HUMAN
|
||||||
| θ value | 0.125558 (rank : 68) | NC score | 0.182645 (rank : 73) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O94842 | Gene names | C14orf92, KIAA0737 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
CN092_MOUSE
|
||||||
| θ value | 0.125558 (rank : 69) | NC score | 0.177645 (rank : 75) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
HM2L1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 70) | NC score | 0.266570 (rank : 69) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9UGU5, O75672, O75673, Q9UMT5 | Gene names | HMG2L1, HMGBCG | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 2-like 1 (Protein HMGBCG). | |||||
|
GLIS1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 71) | NC score | 0.004065 (rank : 156) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K1M4, Q6NZF6, Q8R4Z3 | Gene names | Glis1, Gli5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS1 (GLI-similar 1) (Gli homologous protein 1) (GliH1). | |||||
|
4ET_MOUSE
|
||||||
| θ value | 0.21417 (rank : 72) | NC score | 0.062516 (rank : 92) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EST3, Q9CSS3 | Gene names | Eif4enif1, Clast4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
MAGI2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 73) | NC score | 0.019581 (rank : 134) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 0.279714 (rank : 74) | NC score | 0.045202 (rank : 100) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
SSRP1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 75) | NC score | 0.335652 (rank : 60) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 76) | NC score | 0.301897 (rank : 61) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 0.279714 (rank : 77) | NC score | 0.041092 (rank : 106) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
RBM12_HUMAN
|
||||||
| θ value | 0.365318 (rank : 78) | NC score | 0.046307 (rank : 98) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
UBF1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 79) | NC score | 0.291611 (rank : 62) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
ZN207_HUMAN
|
||||||
| θ value | 0.47712 (rank : 80) | NC score | 0.066583 (rank : 89) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 81) | NC score | 0.067922 (rank : 88) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BAG4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 82) | NC score | 0.041027 (rank : 107) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
CNOT4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 83) | NC score | 0.047251 (rank : 97) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
ITCH_MOUSE
|
||||||
| θ value | 0.62314 (rank : 84) | NC score | 0.009623 (rank : 148) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
PB1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 85) | NC score | 0.193756 (rank : 72) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
4ET_HUMAN
|
||||||
| θ value | 0.813845 (rank : 86) | NC score | 0.057802 (rank : 94) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NRA8, Q8NCF2, Q9H708 | Gene names | EIF4ENIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
ABI2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 87) | NC score | 0.021792 (rank : 130) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NYB9, Q13147, Q13249, Q13801, Q9BV70 | Gene names | ABI2, ARGBPIA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2) (Abl-binding protein 3) (AblBP3) (Arg-binding protein 1) (ArgBP1). | |||||
|
CNOT4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 88) | NC score | 0.047907 (rank : 96) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
HMGB1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 89) | NC score | 0.269755 (rank : 68) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P09429, Q6IBE1 | Gene names | HMGB1, HMG1 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HMGB1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 90) | NC score | 0.269825 (rank : 67) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P63158, P07155, P27109, P27428 | Gene names | Hmgb1, Hmg-1, Hmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 91) | NC score | 0.026924 (rank : 119) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
PMS1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 92) | NC score | 0.163781 (rank : 77) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
TFCP2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 93) | NC score | 0.028606 (rank : 116) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12800, Q12801, Q9UD77 | Gene names | TFCP2, LSF, SEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-globin transcription factor CP2 (Transcription factor LSF) (SAA3 enhancer factor). | |||||
|
TFCP2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 94) | NC score | 0.028616 (rank : 115) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERA0, Q3UGJ4, Q8VC13 | Gene names | Tcfcp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-globin transcription factor CP2. | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 95) | NC score | 0.040951 (rank : 108) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 96) | NC score | 0.045777 (rank : 99) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MYCD_HUMAN
|
||||||
| θ value | 1.38821 (rank : 97) | NC score | 0.023871 (rank : 124) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZQ8, Q8N7Q1 | Gene names | MYOCD, MYCD | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin. | |||||
|
PHLB2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 98) | NC score | 0.009808 (rank : 147) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
ST5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 99) | NC score | 0.020320 (rank : 133) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q924W7, Q78H54, Q8K2P3, Q924W8 | Gene names | St5, Dennd2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (DENN domain-containing protein 2B). | |||||
|
HMGB2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 100) | NC score | 0.289451 (rank : 63) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P26583 | Gene names | HMGB2, HMG2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
IF4G3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 101) | NC score | 0.021993 (rank : 129) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 1.81305 (rank : 102) | NC score | 0.023321 (rank : 126) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
SPT5H_HUMAN
|
||||||
| θ value | 1.81305 (rank : 103) | NC score | 0.044662 (rank : 102) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
SPT5H_MOUSE
|
||||||
| θ value | 1.81305 (rank : 104) | NC score | 0.044791 (rank : 101) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
ABLM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 105) | NC score | 0.018044 (rank : 135) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14639, Q15039, Q68CQ9, Q9BUP1 | Gene names | ABLIM1, ABLIM, KIAA0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1) (Actin-binding double-zinc-finger protein) (LIMAB1) (Limatin). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 106) | NC score | 0.017913 (rank : 136) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 107) | NC score | 0.030068 (rank : 114) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 108) | NC score | 0.050055 (rank : 95) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 2.36792 (rank : 109) | NC score | 0.016799 (rank : 139) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
PRR13_HUMAN
|
||||||
| θ value | 2.36792 (rank : 110) | NC score | 0.101520 (rank : 84) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
RERE_HUMAN
|
||||||
| θ value | 2.36792 (rank : 111) | NC score | 0.062408 (rank : 93) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.044379 (rank : 103) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 113) | NC score | 0.044332 (rank : 104) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
DMRT1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 114) | NC score | 0.022968 (rank : 127) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZ59, Q9QZ37, Q9QZA4 | Gene names | Dmrt1 | |||
|
Domain Architecture |
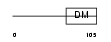
|
|||||
| Description | Doublesex- and mab-3-related transcription factor 1. | |||||
|
EFS_HUMAN
|
||||||
| θ value | 3.0926 (rank : 115) | NC score | 0.024963 (rank : 122) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
ENAH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 116) | NC score | 0.026677 (rank : 120) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
HMGB2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 117) | NC score | 0.270488 (rank : 66) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P30681, Q3UXT1, Q9EQD5 | Gene names | Hmgb2, Hmg2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
IRX2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 118) | NC score | 0.007633 (rank : 152) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BZI1, Q7Z2I7 | Gene names | IRX2, IRXA2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-2 (Iroquois homeobox protein 2) (Homeodomain protein IRXA2). | |||||
|
PCD15_HUMAN
|
||||||
| θ value | 3.0926 (rank : 119) | NC score | 0.004824 (rank : 154) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96QU1, Q96QT8 | Gene names | PCDH15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
RPB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 120) | NC score | 0.036459 (rank : 109) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 121) | NC score | 0.036243 (rank : 111) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
AT11A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.020345 (rank : 132) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98196, Q5VXT2 | Gene names | ATP11A, ATPIH, ATPIS, KIAA1021 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | 0.010981 (rank : 145) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
NELFA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 124) | NC score | 0.022700 (rank : 128) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 125) | NC score | 0.025403 (rank : 121) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SMCE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 126) | NC score | 0.182351 (rank : 74) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
SMCE1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 127) | NC score | 0.171574 (rank : 76) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.024418 (rank : 123) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
DIP2C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.008896 (rank : 149) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2E4 | Gene names | DIP2C, KIAA0934 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog C. | |||||
|
FIGN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.012156 (rank : 142) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
FOXGB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.005141 (rank : 153) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55315 | Gene names | FOXG1B, FKHL1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein G1B (Forkhead-related protein FKHL1) (Transcription factor BF-1) (Brain factor 1) (BF1) (HFK1). | |||||
|
KIF12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.004792 (rank : 155) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96FN5, Q5TBE0 | Gene names | KIF12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF12. | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.041789 (rank : 105) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
SMAD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.007950 (rank : 151) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70340, P70442, Q9CYK6 | Gene names | Smad1, Madh1, Madr1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Mothers-against-DPP-related 1) (mMad1) (Dwarfin-A) (Dwf-A). | |||||
|
TMBI1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | 0.010379 (rank : 146) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BJZ3, Q8CH91, Q99KB6 | Gene names | Tmbim1, Recs1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane BAX inhibitor motif-containing protein 1 (RECS1 protein) (Responsive to centrifugal force and shear stress gene 1 protein). | |||||
|
AT11A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.016998 (rank : 138) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98197 | Gene names | Atp11a | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
IF4G3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.017803 (rank : 137) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
MNT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.023432 (rank : 125) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
RBM12_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.036449 (rank : 110) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.065002 (rank : 91) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RUNX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.011273 (rank : 144) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13950, O14614, O14615, O95181 | Gene names | RUNX2, AML3, CBFA1, OSF2, PEBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
RUNX2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.013882 (rank : 141) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
SGIP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.021042 (rank : 131) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VD37, Q3UFU3, Q3UGA0, Q8BXX4, Q8C034, Q9CXT2 | Gene names | Sgip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
TFAM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.208443 (rank : 71) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P40630, P97894, P97906, Q543I8, Q9DBM9 | Gene names | Tfam, Hmgts | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Testis- specific high mobility group protein) (TS-HMG). | |||||
|
FIP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.014433 (rank : 140) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D824, Q8BWX7, Q99LH0, Q9DBB2 | Gene names | Fip1l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1). | |||||
|
HNF3A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.003516 (rank : 157) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35582, Q61108 | Gene names | Foxa1, Hnf3a, Tcf-3a, Tcf3a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-alpha (HNF-3A) (Forkhead box protein A1). | |||||
|
KCNA5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.002653 (rank : 158) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
LNP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.008843 (rank : 150) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9C0E8, Q2M2V8, Q2YD99, Q658W8, Q8N5V9, Q96MS5 | Gene names | LNP, KIAA1715 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein lunapark. | |||||
|
MYO15_MOUSE
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.011645 (rank : 143) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
SYN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.028033 (rank : 117) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
HMG1X_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.218006 (rank : 70) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UGV6 | Gene names | HMG1L10 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 1-like 10 (HMG-1L10). | |||||
|
HMGB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.280530 (rank : 64) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O15347, O95556, Q6NS40 | Gene names | HMGB3, HMG2A, HMG4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
HMGB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.277949 (rank : 65) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O54879 | Gene names | Hmgb3, Hmg2a, Hmg4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
PRR13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.083322 (rank : 86) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SP100_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.122396 (rank : 82) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P23497, O75450, Q13343, Q96F70, Q96T24, Q96T95, Q9NP33, Q9UE32 | Gene names | SP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein) (Lysp100b). | |||||
|
TFAM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.144263 (rank : 79) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q00059 | Gene names | TFAM, TCF6L2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Mitochondrial transcription factor 1) (MtTF1) (Transcription factor 6-like 2). | |||||
|
TOX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.131128 (rank : 81) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O94900, Q96AV5 | Gene names | TOX, KIAA0808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
TOX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.131356 (rank : 80) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q66JW3, Q8BKH9, Q8BYQ5, Q8R4H0 | Gene names | Tox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
LEF1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 150 | |
| SwissProt Accessions | Q9UJU2, Q9HAZ0 | Gene names | LEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1) (T cell-specific transcription factor 1-alpha) (TCF1-alpha). | |||||
|
LEF1_MOUSE
|
||||||
| NC score | 0.997231 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 141 | |
| SwissProt Accessions | P27782 | Gene names | Lef1, Lef-1 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphoid enhancer-binding factor 1 (LEF-1). | |||||
|
TF7L2_MOUSE
|
||||||
| NC score | 0.971259 (rank : 3) | θ value | 4.32305e-111 (rank : 3) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q924A0, O70574, Q91XP2, Q91XP3, Q91XP4, Q924A1, Q9Z0V3, Q9Z0V4 | Gene names | Tcf7l2, Tcf4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (mTCF-4). | |||||
|
TF7L2_HUMAN
|
||||||
| NC score | 0.963767 (rank : 4) | θ value | 7.14227e-106 (rank : 4) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9NQB0, O00185, Q9NQB1, Q9NQB2, Q9NQB3, Q9NQB4, Q9NQB5, Q9NQB6, Q9NQB7, Q9ULC2 | Gene names | TCF7L2, TCF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (hTCF-4). | |||||
|
TF7L1_HUMAN
|
||||||
| NC score | 0.957551 (rank : 5) | θ value | 2.01746e-92 (rank : 5) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9HCS4, Q53R97, Q6PD70, Q9NP00 | Gene names | TCF7L1, TCF3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF- 3). | |||||
|
TF7L1_MOUSE
|
||||||
| NC score | 0.956415 (rank : 6) | θ value | 4.96919e-91 (rank : 6) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Z1J1, O70450, O70573 | Gene names | Tcf7l1, Tcf3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 1 (HMG box transcription factor 3) (TCF-3) (mTCF-3). | |||||
|
TCF7_HUMAN
|
||||||
| NC score | 0.937600 (rank : 7) | θ value | 6.32992e-62 (rank : 8) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P36402, Q9UKI4 | Gene names | TCF7, TCF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
TCF7_MOUSE
|
||||||
| NC score | 0.930985 (rank : 8) | θ value | 9.41505e-74 (rank : 7) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
SOX30_MOUSE
|
||||||
| NC score | 0.606068 (rank : 9) | θ value | 2.36244e-08 (rank : 16) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SRY_HUMAN
|
||||||
| NC score | 0.605206 (rank : 10) | θ value | 3.77169e-06 (rank : 34) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q05066 | Gene names | SRY, TDF | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
SOX30_HUMAN
|
||||||
| NC score | 0.605070 (rank : 11) | θ value | 2.36244e-08 (rank : 15) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SOX12_MOUSE
|
||||||
| NC score | 0.603747 (rank : 12) | θ value | 2.21117e-06 (rank : 27) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q04890, P70417, Q6NXL2 | Gene names | Sox12, Sox-12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SOX-12 protein. | |||||
|
SOX12_HUMAN
|
||||||
| NC score | 0.602293 (rank : 13) | θ value | 2.21117e-06 (rank : 26) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O15370, Q9NUD4 | Gene names | SOX12, SOX22 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-12 protein (SOX-22 protein). | |||||
|
SOX7_MOUSE
|
||||||
| NC score | 0.597534 (rank : 14) | θ value | 1.53129e-07 (rank : 20) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P40646, Q9R1T6 | Gene names | Sox7, Sox-7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7 (mSOX7). | |||||
|
SOX7_HUMAN
|
||||||
| NC score | 0.593599 (rank : 15) | θ value | 1.53129e-07 (rank : 19) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
SRY_MOUSE
|
||||||
| NC score | 0.593058 (rank : 16) | θ value | 7.59969e-07 (rank : 21) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q05738 | Gene names | Sry, Tdf, Tdy | |||
|
Domain Architecture |

|
|||||
| Description | Sex-determining region Y protein (Testis-determining factor). | |||||
|
SOX14_MOUSE
|
||||||
| NC score | 0.588035 (rank : 17) | θ value | 2.21117e-06 (rank : 29) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q04892, Q9JLC9, Q9JLD0, Q9QXU4 | Gene names | Sox14, Sox-14 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX14_HUMAN
|
||||||
| NC score | 0.587959 (rank : 18) | θ value | 2.21117e-06 (rank : 28) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O95416, Q3KPH7 | Gene names | SOX14, SOX28 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
SOX11_HUMAN
|
||||||
| NC score | 0.587319 (rank : 19) | θ value | 2.88788e-06 (rank : 30) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P35716 | Gene names | SOX11 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX2_HUMAN
|
||||||
| NC score | 0.586402 (rank : 20) | θ value | 9.92553e-07 (rank : 22) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P48431, Q14537 | Gene names | SOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX2_MOUSE
|
||||||
| NC score | 0.586050 (rank : 21) | θ value | 9.92553e-07 (rank : 23) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P48432 | Gene names | Sox2, Sox-2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-2. | |||||
|
SOX11_MOUSE
|
||||||
| NC score | 0.585804 (rank : 22) | θ value | 2.88788e-06 (rank : 31) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q7M6Y2, O35178, O89036, Q04889, Q80XF0 | Gene names | Sox11, Sox-11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-11. | |||||
|
SOX21_HUMAN
|
||||||
| NC score | 0.585728 (rank : 23) | θ value | 3.77169e-06 (rank : 32) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Y651, P35715, Q15504 | Gene names | SOX21, SOX25, SOXA | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-21 (SOX-A). | |||||
|
SOX21_MOUSE
|
||||||
| NC score | 0.585699 (rank : 24) | θ value | 3.77169e-06 (rank : 33) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q811W0, Q8VH35 | Gene names | Sox21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-21. | |||||
|
SOX6_HUMAN
|
||||||
| NC score | 0.584684 (rank : 25) | θ value | 1.63604e-09 (rank : 11) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P35712, Q9BXQ3, Q9BXQ4, Q9BXQ5, Q9H0I8 | Gene names | SOX6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6. | |||||
|
SOX6_MOUSE
|
||||||
| NC score | 0.584027 (rank : 26) | θ value | 1.63604e-09 (rank : 12) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P40645, Q62250, Q9QWS5 | Gene names | Sox6, Sox-6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6 (SOX-LZ). | |||||
|
SOX5_MOUSE
|
||||||
| NC score | 0.581667 (rank : 27) | θ value | 1.80886e-08 (rank : 14) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P35710, O88184, O89018 | Gene names | Sox5, Sox-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX8_HUMAN
|
||||||
| NC score | 0.581349 (rank : 28) | θ value | 9.92553e-07 (rank : 24) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P57073, Q9NZW2 | Gene names | SOX8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX5_HUMAN
|
||||||
| NC score | 0.579940 (rank : 29) | θ value | 1.80886e-08 (rank : 13) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX4_MOUSE
|
||||||
| NC score | 0.579049 (rank : 30) | θ value | 1.09739e-05 (rank : 37) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q06831 | Gene names | Sox4, Sox-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX8_MOUSE
|
||||||
| NC score | 0.578047 (rank : 31) | θ value | 9.92553e-07 (rank : 25) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q04886 | Gene names | Sox8, Sox-8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
SOX4_HUMAN
|
||||||
| NC score | 0.577903 (rank : 32) | θ value | 1.87187e-05 (rank : 44) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q06945 | Gene names | SOX4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SOX18_MOUSE
|
||||||
| NC score | 0.576156 (rank : 33) | θ value | 3.19293e-05 (rank : 45) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P43680, Q9EQ73 | Gene names | Sox18, Sox-18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
SOX17_HUMAN
|
||||||
| NC score | 0.575720 (rank : 34) | θ value | 8.40245e-06 (rank : 35) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9H6I2 | Gene names | SOX17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX17_MOUSE
|
||||||
| NC score | 0.575438 (rank : 35) | θ value | 8.40245e-06 (rank : 36) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q61473, Q61472, Q62248 | Gene names | Sox17, Sox-17 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-17. | |||||
|
SOX13_HUMAN
|
||||||
| NC score | 0.575117 (rank : 36) | θ value | 4.0297e-08 (rank : 17) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.571257 (rank : 37) | θ value | 4.0297e-08 (rank : 18) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SOX1_HUMAN
|
||||||
| NC score | 0.569653 (rank : 38) | θ value | 1.87187e-05 (rank : 42) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O00570 | Gene names | SOX1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX18_HUMAN
|
||||||
| NC score | 0.569386 (rank : 39) | θ value | 7.1131e-05 (rank : 50) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P35713, Q9NPH8 | Gene names | SOX18 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-18. | |||||
|
SOX1_MOUSE
|
||||||
| NC score | 0.569105 (rank : 40) | θ value | 1.87187e-05 (rank : 43) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P53783 | Gene names | Sox1, Sox-1 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-1 protein. | |||||
|
SOX15_MOUSE
|
||||||
| NC score | 0.566198 (rank : 41) | θ value | 5.44631e-05 (rank : 46) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P43267, O70204, P70418, Q62246, Q91V00, Q91V43, Q920T1, Q9JLG2 | Gene names | Sox15, Sox-15 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein. | |||||
|
SOX15_HUMAN
|
||||||
| NC score | 0.562293 (rank : 42) | θ value | 7.1131e-05 (rank : 49) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | O60248, P35717, Q9Y6W7 | Gene names | SOX15, SOX12, SOX20, SOX26, SOX27 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein (SOX-20 protein) (SOX-12 protein). | |||||
|
SOX9_MOUSE
|
||||||
| NC score | 0.561262 (rank : 43) | θ value | 1.43324e-05 (rank : 40) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q04887, Q8C7L2, Q91ZK2, Q99KQ0 | Gene names | Sox9, Sox-9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX9_HUMAN
|
||||||
| NC score | 0.560619 (rank : 44) | θ value | 1.43324e-05 (rank : 39) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SOX10_HUMAN
|
||||||
| NC score | 0.559922 (rank : 45) | θ value | 1.43324e-05 (rank : 38) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
SOX3_MOUSE
|
||||||
| NC score | 0.559002 (rank : 46) | θ value | 5.44631e-05 (rank : 48) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P53784 | Gene names | Sox3, Sox-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SOX10_MOUSE
|
||||||
| NC score | 0.557793 (rank : 47) | θ value | 1.87187e-05 (rank : 41) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q04888, O08518, O09141, O54856, P70416 | Gene names | Sox10, Sox-10, Sox21 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10 (SOX-21) (Transcription factor SOX-M). | |||||
|
SOX3_HUMAN
|
||||||
| NC score | 0.555964 (rank : 48) | θ value | 5.44631e-05 (rank : 47) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P41225, P35714, Q9NP49 | Gene names | SOX3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.554293 (rank : 49) | θ value | 5.62301e-10 (rank : 10) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.527309 (rank : 50) | θ value | 5.62301e-10 (rank : 9) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
HBP1_MOUSE
|
||||||
| NC score | 0.508913 (rank : 51) | θ value | 0.000461057 (rank : 54) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8R316, Q3V0I4, Q8BUS3, Q8C199 | Gene names | Hbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
HBP1_HUMAN
|
||||||
| NC score | 0.507406 (rank : 52) | θ value | 0.000461057 (rank : 53) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O60381, Q8TBM1, Q8TE93, Q96AJ2 | Gene names | HBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box-containing protein 1 (HMG box transcription factor 1) (High mobility group box transcription factor 1). | |||||
|
BBX_MOUSE
|
||||||
| NC score | 0.487445 (rank : 53) | θ value | 0.000270298 (rank : 52) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8VBW5, Q3TZK1, Q6NXY8, Q6PEU3, Q8BQJ7, Q8C7E0, Q8CDQ0, Q8CDV1, Q8VI48, Q8VI49, Q8VI50, Q9CS94 | Gene names | Bbx, Hbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
BBX_HUMAN
|
||||||
| NC score | 0.484139 (rank : 54) | θ value | 0.000270298 (rank : 51) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8WY36, Q2TAJ1, Q7L3J8, Q7LBY8, Q8NDB0, Q8WY35, Q9H0J6 | Gene names | BBX, HBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
HM20B_HUMAN
|
||||||
| NC score | 0.364272 (rank : 55) | θ value | 0.0330416 (rank : 62) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9P0W2, Q6IBP8, Q8NBD5, Q9HD21, Q9Y491, Q9Y4A2 | Gene names | HMG20B, BRAF35, HMGX2, SMARCE1R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35) (Sox-like transcriptional factor). | |||||
|
HM20B_MOUSE
|
||||||
| NC score | 0.360874 (rank : 56) | θ value | 0.0431538 (rank : 63) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Z104, Q9NSF7, Q9NSF8 | Gene names | Hmg20b, Braf35, Hmgx2, Smarce1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1-related (SMARCE1-related protein) (HMG domain protein HMG20B) (Structural DNA-binding protein BRAF35) (BRCA2- associated factor 35). | |||||
|
HM20A_HUMAN
|
||||||
| NC score | 0.356240 (rank : 57) | θ value | 0.0148317 (rank : 57) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9NP66, Q53G31, Q9NSF6 | Gene names | HMG20A, HMGX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (HMG domain-containing protein HMGX1). | |||||
|
HM20A_MOUSE
|
||||||
| NC score | 0.355349 (rank : 58) | θ value | 0.0148317 (rank : 58) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9DC33, Q3LSF9, Q6NV87, Q8BSK1, Q8C3C1, Q8CAA0, Q9CYG2 | Gene names | Hmg20a, Ibraf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (iBRAF) (Inhibitor of BRAF35) (HMG domain protein HMGX1). | |||||
|
SSRP1_MOUSE
|
||||||
| NC score | 0.339519 (rank : 59) | θ value | 0.0961366 (rank : 67) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
SSRP1_HUMAN
|
||||||
| NC score | 0.335652 (rank : 60) | θ value | 0.279714 (rank : 75) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.301897 (rank : 61) | θ value | 0.279714 (rank : 76) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
UBF1_MOUSE
|
||||||
| NC score | 0.291611 (rank : 62) | θ value | 0.47712 (rank : 79) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
HMGB2_HUMAN
|
||||||
| NC score | 0.289451 (rank : 63) | θ value | 1.81305 (rank : 100) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P26583 | Gene names | HMGB2, HMG2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
HMGB3_HUMAN
|
||||||
| NC score | 0.280530 (rank : 64) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O15347, O95556, Q6NS40 | Gene names | HMGB3, HMG2A, HMG4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
HMGB3_MOUSE
|
||||||
| NC score | 0.277949 (rank : 65) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O54879 | Gene names | Hmgb3, Hmg2a, Hmg4 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B3 (High mobility group protein 4) (HMG-4) (High mobility group protein 2a) (HMG-2a). | |||||
|
HMGB2_MOUSE
|
||||||
| NC score | 0.270488 (rank : 66) | θ value | 3.0926 (rank : 117) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P30681, Q3UXT1, Q9EQD5 | Gene names | Hmgb2, Hmg2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
HMGB1_MOUSE
|
||||||
| NC score | 0.269825 (rank : 67) | θ value | 0.813845 (rank : 90) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P63158, P07155, P27109, P27428 | Gene names | Hmgb1, Hmg-1, Hmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HMGB1_HUMAN
|
||||||
| NC score | 0.269755 (rank : 68) | θ value | 0.813845 (rank : 89) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P09429, Q6IBE1 | Gene names | HMGB1, HMG1 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B1 (High mobility group protein 1) (HMG- 1). | |||||
|
HM2L1_HUMAN
|
||||||
| NC score | 0.266570 (rank : 69) | θ value | 0.125558 (rank : 70) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9UGU5, O75672, O75673, Q9UMT5 | Gene names | HMG2L1, HMGBCG | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 2-like 1 (Protein HMGBCG). | |||||
|
HMG1X_HUMAN
|
||||||
| NC score | 0.218006 (rank : 70) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UGV6 | Gene names | HMG1L10 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein 1-like 10 (HMG-1L10). | |||||
|
TFAM_MOUSE
|
||||||
| NC score | 0.208443 (rank : 71) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P40630, P97894, P97906, Q543I8, Q9DBM9 | Gene names | Tfam, Hmgts | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Testis- specific high mobility group protein) (TS-HMG). | |||||
|
PB1_HUMAN
|
||||||
| NC score | 0.193756 (rank : 72) | θ value | 0.62314 (rank : 85) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
CN092_HUMAN
|
||||||
| NC score | 0.182645 (rank : 73) | θ value | 0.125558 (rank : 68) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O94842 | Gene names | C14orf92, KIAA0737 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
SMCE1_HUMAN
|
||||||
| NC score | 0.182351 (rank : 74) | θ value | 4.03905 (rank : 126) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
CN092_MOUSE
|
||||||
| NC score | 0.177645 (rank : 75) | θ value | 0.125558 (rank : 69) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
SMCE1_MOUSE
|
||||||
| NC score | 0.171574 (rank : 76) | θ value | 4.03905 (rank : 127) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
PMS1_HUMAN
|
||||||
| NC score | 0.163781 (rank : 77) | θ value | 0.813845 (rank : 92) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
GCX1_HUMAN
|
||||||
| NC score | 0.151385 (rank : 78) | θ value | 0.0736092 (rank : 64) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96NM4, Q5TE33, Q5TE34, Q5TE35, Q96IC9, Q9BQN5 | Gene names | GCX1, C20orf100 | |||
|
Domain Architecture |

|
|||||
| Description | Granulosa cell HMG box protein 1 (GCX-1). | |||||
|
TFAM_HUMAN
|
||||||
| NC score | 0.144263 (rank : 79) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q00059 | Gene names | TFAM, TCF6L2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor A, mitochondrial precursor (mtTFA) (Mitochondrial transcription factor 1) (MtTF1) (Transcription factor 6-like 2). | |||||
|
TOX_MOUSE
|
||||||
| NC score | 0.131356 (rank : 80) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q66JW3, Q8BKH9, Q8BYQ5, Q8R4H0 | Gene names | Tox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
TOX_HUMAN
|
||||||
| NC score | 0.131128 (rank : 81) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O94900, Q96AV5 | Gene names | TOX, KIAA0808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
SP100_HUMAN
|
||||||
| NC score | 0.122396 (rank : 82) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P23497, O75450, Q13343, Q96F70, Q96T24, Q96T95, Q9NP33, Q9UE32 | Gene names | SP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein) (Lysp100b). | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.111594 (rank : 83) | θ value | 0.0148317 (rank : 60) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.101520 (rank : 84) | θ value | 2.36792 (rank : 110) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.095937 (rank : 85) | θ value | 0.0961366 (rank : 66) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
PRR13_MOUSE
|
||||||
| NC score | 0.083322 (rank : 86) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.069688 (rank : 87) | θ value | 0.00509761 (rank : 55) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.067922 (rank : 88) | θ value | 0.62314 (rank : 81) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.066583 (rank : 89) | θ value | 0.47712 (rank : 80) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
MEF2B_MOUSE
|
||||||
| NC score | 0.066142 (rank : 90) | θ value | 0.0148317 (rank : 59) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.065002 (rank : 91) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
4ET_MOUSE
|
||||||
| NC score | 0.062516 (rank : 92) | θ value | 0.21417 (rank : 72) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EST3, Q9CSS3 | Gene names | Eif4enif1, Clast4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.062408 (rank : 93) | θ value | 2.36792 (rank : 111) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
4ET_HUMAN
|
||||||
| NC score | 0.057802 (rank : 94) | θ value | 0.813845 (rank : 86) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NRA8, Q8NCF2, Q9H708 | Gene names | EIF4ENIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.050055 (rank : 95) | θ value | 2.36792 (rank : 108) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
CNOT4_MOUSE
|
||||||
| NC score | 0.047907 (rank : 96) | θ value | 0.813845 (rank : 88) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
CNOT4_HUMAN
|
||||||
| NC score | 0.047251 (rank : 97) | θ value | 0.62314 (rank : 83) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
RBM12_HUMAN
|
||||||
| NC score | 0.046307 (rank : 98) | θ value | 0.365318 (rank : 78) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.045777 (rank : 99) | θ value | 1.38821 (rank : 96) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.045202 (rank : 100) | θ value | 0.279714 (rank : 74) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
SPT5H_MOUSE
|
||||||
| NC score | 0.044791 (rank : 101) | θ value | 1.81305 (rank : 104) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
SPT5H_HUMAN
|
||||||
| NC score | 0.044662 (rank : 102) | θ value | 1.81305 (rank : 103) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.044379 (rank : 103) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.044332 (rank : 104) | θ value | 3.0926 (rank : 113) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.041789 (rank : 105) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.041092 (rank : 106) | θ value | 0.279714 (rank : 77) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
BAG4_HUMAN
|
||||||
| NC score | 0.041027 (rank : 107) | θ value | 0.62314 (rank : 82) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.040951 (rank : 108) | θ value | 1.06291 (rank : 95) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.036459 (rank : 109) | θ value | 3.0926 (rank : 120) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RBM12_MOUSE
|
||||||
| NC score | 0.036449 (rank : 110) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.036243 (rank : 111) | θ value | 3.0926 (rank : 121) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
ANX11_HUMAN
|
||||||
| NC score | 0.034493 (rank : 112) | θ value | 0.0148317 (rank : 56) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P50995 | Gene names | ANXA11, ANX11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50) (56 kDa autoantigen). | |||||
|
HXB3_HUMAN
|
||||||
| NC score | 0.031731 (rank : 113) | θ value | 0.0252991 (rank : 61) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P14651, O95615, P17484 | Gene names | HOXB3, HOX2G | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2G) (Hox-2.7). | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.030068 (rank : 114) | θ value | 2.36792 (rank : 107) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
TFCP2_MOUSE
|
||||||
| NC score | 0.028616 (rank : 115) | θ value | 0.813845 (rank : 94) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERA0, Q3UGJ4, Q8VC13 | Gene names | Tcfcp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-globin transcription factor CP2. | |||||
|
TFCP2_HUMAN
|
||||||
| NC score | 0.028606 (rank : 116) | θ value | 0.813845 (rank : 93) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12800, Q12801, Q9UD77 | Gene names | TFCP2, LSF, SEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-globin transcription factor CP2 (Transcription factor LSF) (SAA3 enhancer factor). | |||||
|
SYN1_HUMAN
|
||||||
| NC score | 0.028033 (rank : 117) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
HXB3_MOUSE
|
||||||
| NC score | 0.027214 (rank : 118) | θ value | 0.0736092 (rank : 65) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P09026, P10285, Q4PJ06, Q61680 | Gene names | Hoxb3, Hox-2.7, Hoxb-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2.7) (Homeobox protein MH-23). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.026924 (rank : 119) | θ value | 0.813845 (rank : 91) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
ENAH_HUMAN
|
||||||
| NC score | 0.026677 (rank : 120) | θ value | 3.0926 (rank : 116) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.025403 (rank : 121) | θ value | 4.03905 (rank : 125) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
EFS_HUMAN
|
||||||
| NC score | 0.024963 (rank : 122) | θ value | 3.0926 (rank : 115) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.024418 (rank : 123) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
MYCD_HUMAN
|
||||||
| NC score | 0.023871 (rank : 124) | θ value | 1.38821 (rank : 97) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZQ8, Q8N7Q1 | Gene names | MYOCD, MYCD | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin. | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.023432 (rank : 125) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.023321 (rank : 126) | θ value | 1.81305 (rank : 102) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
DMRT1_MOUSE
|
||||||
| NC score | 0.022968 (rank : 127) | θ value | 3.0926 (rank : 114) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZ59, Q9QZ37, Q9QZA4 | Gene names | Dmrt1 | |||
|
Domain Architecture |

|
|||||
| Description | Doublesex- and mab-3-related transcription factor 1. | |||||
|
NELFA_MOUSE
|
||||||
| NC score | 0.022700 (rank : 128) | θ value | 4.03905 (rank : 124) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
IF4G3_HUMAN
|
||||||
| NC score | 0.021993 (rank : 129) | θ value | 1.81305 (rank : 101) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
ABI2_HUMAN
|
||||||
| NC score | 0.021792 (rank : 130) | θ value | 0.813845 (rank : 87) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NYB9, Q13147, Q13249, Q13801, Q9BV70 | Gene names | ABI2, ARGBPIA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2) (Abl-binding protein 3) (AblBP3) (Arg-binding protein 1) (ArgBP1). | |||||
|
SGIP1_MOUSE
|
||||||
| NC score | 0.021042 (rank : 131) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VD37, Q3UFU3, Q3UGA0, Q8BXX4, Q8C034, Q9CXT2 | Gene names | Sgip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
AT11A_HUMAN
|
||||||
| NC score | 0.020345 (rank : 132) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98196, Q5VXT2 | Gene names | ATP11A, ATPIH, ATPIS, KIAA1021 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
ST5_MOUSE
|
||||||
| NC score | 0.020320 (rank : 133) | θ value | 1.38821 (rank : 99) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q924W7, Q78H54, Q8K2P3, Q924W8 | Gene names | St5, Dennd2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (DENN domain-containing protein 2B). | |||||
|
MAGI2_HUMAN
|
||||||
| NC score | 0.019581 (rank : 134) | θ value | 0.21417 (rank : 73) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
ABLM1_HUMAN
|
||||||
| NC score | 0.018044 (rank : 135) | θ value | 2.36792 (rank : 105) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14639, Q15039, Q68CQ9, Q9BUP1 | Gene names | ABLIM1, ABLIM, KIAA0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1) (Actin-binding double-zinc-finger protein) (LIMAB1) (Limatin). | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.017913 (rank : 136) | θ value | 2.36792 (rank : 106) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
IF4G3_MOUSE
|
||||||
| NC score | 0.017803 (rank : 137) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
AT11A_MOUSE
|
||||||
| NC score | 0.016998 (rank : 138) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98197 | Gene names | Atp11a | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IH (EC 3.6.3.1) (ATPase class I type 11A) (ATPase IS). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.016799 (rank : 139) | θ value | 2.36792 (rank : 109) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
FIP1_MOUSE
|
||||||
| NC score | 0.014433 (rank : 140) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D824, Q8BWX7, Q99LH0, Q9DBB2 | Gene names | Fip1l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1). | |||||
|
RUNX2_MOUSE
|
||||||
| NC score | 0.013882 (rank : 141) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
FIGN_MOUSE
|
||||||
| NC score | 0.012156 (rank : 142) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.011645 (rank : 143) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
RUNX2_HUMAN
|
||||||
| NC score | 0.011273 (rank : 144) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13950, O14614, O14615, O95181 | Gene names | RUNX2, AML3, CBFA1, OSF2, PEBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.010981 (rank : 145) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
TMBI1_MOUSE
|
||||||
| NC score | 0.010379 (rank : 146) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BJZ3, Q8CH91, Q99KB6 | Gene names | Tmbim1, Recs1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane BAX inhibitor motif-containing protein 1 (RECS1 protein) (Responsive to centrifugal force and shear stress gene 1 protein). | |||||
|
PHLB2_HUMAN
|
||||||
| NC score | 0.009808 (rank : 147) | θ value | 1.38821 (rank : 98) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
ITCH_MOUSE
|
||||||
| NC score | 0.009623 (rank : 148) | θ value | 0.62314 (rank : 84) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
DIP2C_HUMAN
|
||||||
| NC score | 0.008896 (rank : 149) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2E4 | Gene names | DIP2C, KIAA0934 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog C. | |||||
|
LNP_HUMAN
|
||||||
| NC score | 0.008843 (rank : 150) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9C0E8, Q2M2V8, Q2YD99, Q658W8, Q8N5V9, Q96MS5 | Gene names | LNP, KIAA1715 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein lunapark. | |||||
|
SMAD1_MOUSE
|
||||||
| NC score | 0.007950 (rank : 151) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70340, P70442, Q9CYK6 | Gene names | Smad1, Madh1, Madr1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Mothers-against-DPP-related 1) (mMad1) (Dwarfin-A) (Dwf-A). | |||||
|
IRX2_HUMAN
|
||||||
| NC score | 0.007633 (rank : 152) | θ value | 3.0926 (rank : 118) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BZI1, Q7Z2I7 | Gene names | IRX2, IRXA2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-2 (Iroquois homeobox protein 2) (Homeodomain protein IRXA2). | |||||
|
FOXGB_HUMAN
|
||||||
| NC score | 0.005141 (rank : 153) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55315 | Gene names | FOXG1B, FKHL1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein G1B (Forkhead-related protein FKHL1) (Transcription factor BF-1) (Brain factor 1) (BF1) (HFK1). | |||||
|
PCD15_HUMAN
|
||||||
| NC score | 0.004824 (rank : 154) | θ value | 3.0926 (rank : 119) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96QU1, Q96QT8 | Gene names | PCDH15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
KIF12_HUMAN
|
||||||
| NC score | 0.004792 (rank : 155) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96FN5, Q5TBE0 | Gene names | KIF12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF12. | |||||
|
GLIS1_MOUSE
|
||||||
| NC score | 0.004065 (rank : 156) | θ value | 0.163984 (rank : 71) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K1M4, Q6NZF6, Q8R4Z3 | Gene names | Glis1, Gli5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS1 (GLI-similar 1) (Gli homologous protein 1) (GliH1). | |||||
|
HNF3A_MOUSE
|
||||||
| NC score | 0.003516 (rank : 157) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35582, Q61108 | Gene names | Foxa1, Hnf3a, Tcf-3a, Tcf3a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-alpha (HNF-3A) (Forkhead box protein A1). | |||||
|
KCNA5_HUMAN
|
||||||
| NC score | 0.002653 (rank : 158) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||