Please be patient as the page loads
|
GATA6_MOUSE
|
||||||
| SwissProt Accessions | Q61169, P97729, Q9QZK3 | Gene names | Gata6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GATA6_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q61169, P97729, Q9QZK3 | Gene names | Gata6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GATA6_HUMAN
|
||||||
| θ value | 1.27474e-179 (rank : 2) | NC score | 0.992422 (rank : 2) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q92908, P78327 | Gene names | GATA6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GATA5_MOUSE
|
||||||
| θ value | 2.55803e-79 (rank : 3) | NC score | 0.974188 (rank : 3) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97489 | Gene names | Gata5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
GATA5_HUMAN
|
||||||
| θ value | 5.69867e-79 (rank : 4) | NC score | 0.972434 (rank : 4) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BWX5 | Gene names | GATA5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
GATA4_HUMAN
|
||||||
| θ value | 2.24094e-75 (rank : 5) | NC score | 0.972302 (rank : 5) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P43694 | Gene names | GATA4 | |||
|
Domain Architecture |
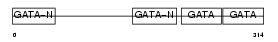
|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GATA4_MOUSE
|
||||||
| θ value | 8.51556e-75 (rank : 6) | NC score | 0.970254 (rank : 6) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q08369, P97491, Q9QZK4 | Gene names | Gata4, Gata-4 | |||
|
Domain Architecture |
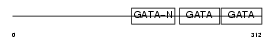
|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GATA3_MOUSE
|
||||||
| θ value | 1.32293e-51 (rank : 7) | NC score | 0.932749 (rank : 9) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P23772 | Gene names | Gata3, Gata-3 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-acting T-cell-specific transcription factor GATA-3 (GATA-binding factor 3). | |||||
|
GATA3_HUMAN
|
||||||
| θ value | 2.94719e-51 (rank : 8) | NC score | 0.931318 (rank : 10) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P23771, Q96J16 | Gene names | GATA3 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-acting T-cell-specific transcription factor GATA-3 (GATA-binding factor 3). | |||||
|
GATA2_HUMAN
|
||||||
| θ value | 3.2585e-50 (rank : 9) | NC score | 0.924965 (rank : 11) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P23769, Q9BUJ6 | Gene names | GATA2 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial transcription factor GATA-2 (GATA-binding protein 2). | |||||
|
GATA1_MOUSE
|
||||||
| θ value | 1.23823e-49 (rank : 10) | NC score | 0.948163 (rank : 7) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17679 | Gene names | Gata1, Gf-1 | |||
|
Domain Architecture |

|
|||||
| Description | Erythroid transcription factor (GATA-binding factor 1) (GATA-1) (Eryf1) (GF-1) (NF-E1 DNA-binding protein). | |||||
|
GATA2_MOUSE
|
||||||
| θ value | 1.23823e-49 (rank : 11) | NC score | 0.922369 (rank : 12) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O09100 | Gene names | Gata2 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial transcription factor GATA-2 (GATA-binding protein 2). | |||||
|
GATA1_HUMAN
|
||||||
| θ value | 6.14528e-49 (rank : 12) | NC score | 0.945371 (rank : 8) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P15976, Q96GB8 | Gene names | GATA1, ERYF1, GF1 | |||
|
Domain Architecture |

|
|||||
| Description | Erythroid transcription factor (GATA-binding factor 1) (GATA-1) (Eryf1) (GF-1) (NF-E1 DNA-binding protein). | |||||
|
TRPS1_MOUSE
|
||||||
| θ value | 4.16044e-13 (rank : 13) | NC score | 0.472515 (rank : 14) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
TRPS1_HUMAN
|
||||||
| θ value | 1.2105e-12 (rank : 14) | NC score | 0.478435 (rank : 13) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UHF7, Q9NWE1, Q9UHH6 | Gene names | TRPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1 (Zinc finger protein GC79) (Tricho-rhino-phalangeal syndrome type I protein). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 15) | NC score | 0.045258 (rank : 19) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.042553 (rank : 21) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.039465 (rank : 23) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
HD_HUMAN
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.047747 (rank : 18) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.025811 (rank : 29) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
HD_MOUSE
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.049329 (rank : 16) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
ZN533_HUMAN
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.045001 (rank : 20) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q569K4, Q49A04, Q6ZMZ7, Q8IY01, Q8N8H2, Q96DK4 | Gene names | ZNF533 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 533. | |||||
|
TSH3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.015059 (rank : 45) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q63HK5, Q9H0G6, Q9P254 | Gene names | TSHZ3, KIAA1474, TSH3, ZNF537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
CAN7_MOUSE
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.055007 (rank : 15) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R1S8, Q9Z0P9 | Gene names | Capn7, Palbh | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
MTA2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.038937 (rank : 24) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O94776, Q9UQB5 | Gene names | MTA2, MTA1L1, PID | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex). | |||||
|
MTA2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.038932 (rank : 25) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R190, Q3TVT8 | Gene names | Mta2, Mta1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.016775 (rank : 43) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.021212 (rank : 32) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
CO4A2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.008530 (rank : 54) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.016134 (rank : 44) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.004418 (rank : 61) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
ZN443_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.001702 (rank : 70) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2A4 | Gene names | ZNF443 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 443 (Krueppel-type zinc finger protein ZK1). | |||||
|
CAN7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.048044 (rank : 17) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6W3 | Gene names | CAPN7, PALBH | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
CDSN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.024073 (rank : 30) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15517, O43509, Q5SQ85, Q5STD2, Q7LA70, Q7LA71, Q86Z04, Q8IZU4, Q8IZU5, Q8IZU6, Q8N5P3, Q95IF9, Q9NP52, Q9NPE0, Q9NPG5, Q9NRH4, Q9NRH5, Q9NRH6, Q9NRH7, Q9NRH8, Q9UBH8, Q9UIN6, Q9UIN7, Q9UIN8, Q9UIN9, Q9UIP0 | Gene names | CDSN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor (S protein). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.011173 (rank : 48) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
ZN306_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.001854 (rank : 68) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BRR0, Q5VXH3, Q92972, Q9H4T3 | Gene names | ZNF306, ZFP47, ZSCAN13 | |||
|
Domain Architecture |
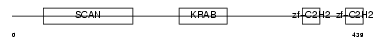
|
|||||
| Description | Zinc finger protein 306 (Zinc finger protein 47 homolog) (Zfp-47) (Zf47) (Zinc finger and SCAN domain-containing protein 13). | |||||
|
ZN533_MOUSE
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.034840 (rank : 26) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BXJ8, Q8BWQ7 | Gene names | Znf533, Zfp533 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 533. | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.008781 (rank : 52) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CIR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.033832 (rank : 27) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
OTX1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.008631 (rank : 53) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P32242, Q53TG6 | Gene names | OTX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein OTX1. | |||||
|
ZN442_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.001523 (rank : 71) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H7R0 | Gene names | ZNF442 | |||
|
Domain Architecture |
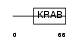
|
|||||
| Description | Zinc finger protein 442. | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.020646 (rank : 33) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
GBRG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.003341 (rank : 63) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0Y8 | Gene names | Gabrg1 | |||
|
Domain Architecture |
|
|||||
| Description | Gamma-aminobutyric-acid receptor subunit gamma-1 precursor (GABA(A) receptor subunit gamma-1). | |||||
|
OTX1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.007241 (rank : 58) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P80205 | Gene names | Otx1, Otx-1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein OTX1. | |||||
|
PININ_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.013551 (rank : 46) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.016898 (rank : 41) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
ZCH10_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.027322 (rank : 28) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
ZN11B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.003776 (rank : 62) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 802 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q06732, Q86XY8, Q8NDW3 | Gene names | ZNF11B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 11B. | |||||
|
ZN672_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.001963 (rank : 67) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99LH4, Q8BIR7 | Gene names | Znf672, Zfp672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 672. | |||||
|
MEF2B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.017722 (rank : 38) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
MTMR5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.008826 (rank : 51) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.018995 (rank : 36) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NP1L3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.008862 (rank : 50) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.019846 (rank : 34) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.019764 (rank : 35) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TAXB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.002349 (rank : 65) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
ZCH10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.041572 (rank : 22) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.011737 (rank : 47) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
RNPS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.017110 (rank : 39) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.017110 (rank : 40) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
RPA2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.010772 (rank : 49) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70700 | Gene names | Polr1b, Rpa2, Rpo1-2 | |||
|
Domain Architecture |
|
|||||
| Description | DNA-directed RNA polymerase I 135 kDa polypeptide (EC 2.7.7.6) (RNA polymerase I subunit 2) (RPA135). | |||||
|
TNC6B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.017921 (rank : 37) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
AKAP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.007764 (rank : 55) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.007586 (rank : 57) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.016802 (rank : 42) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ERR2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.001109 (rank : 72) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61539, O09067, O09146 | Gene names | Esrrb, Err-2, Err2, Nr3b2 | |||
|
Domain Architecture |

|
|||||
| Description | Steroid hormone receptor ERR2 (Estrogen-related receptor, beta) (ERR- beta) (Estrogen receptor-like 2). | |||||
|
HOME1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.002526 (rank : 64) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86YM7, O96003, Q86YM5 | Gene names | HOMER1, SYN47 | |||
|
Domain Architecture |
|
|||||
| Description | Homer protein homolog 1. | |||||
|
HRX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.006906 (rank : 59) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
MTSS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.023427 (rank : 31) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
NF2L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.007761 (rank : 56) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14494, Q12877, Q96FN6 | Gene names | NFE2L1, HBZ17, NRF1, TCF11 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1) (Transcription factor 11) (Transcription factor HBZ17) (Transcription factor LCR-F1) (Locus control region-factor 1). | |||||
|
SOX14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.006174 (rank : 60) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95416, Q3KPH7 | Gene names | SOX14, SOX28 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
ULK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | -0.002184 (rank : 73) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 979 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYT8, O75119 | Gene names | ULK2, KIAA0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2). | |||||
|
ZN509_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.001740 (rank : 69) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ZSB9, Q59FJ4, Q5EBN0, Q8TB80 | Gene names | ZNF509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
ZN509_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.002079 (rank : 66) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
GATA6_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q61169, P97729, Q9QZK3 | Gene names | Gata6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GATA6_HUMAN
|
||||||
| NC score | 0.992422 (rank : 2) | θ value | 1.27474e-179 (rank : 2) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q92908, P78327 | Gene names | GATA6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GATA5_MOUSE
|
||||||
| NC score | 0.974188 (rank : 3) | θ value | 2.55803e-79 (rank : 3) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97489 | Gene names | Gata5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
GATA5_HUMAN
|
||||||
| NC score | 0.972434 (rank : 4) | θ value | 5.69867e-79 (rank : 4) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BWX5 | Gene names | GATA5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-5 (GATA-binding factor 5). | |||||
|
GATA4_HUMAN
|
||||||
| NC score | 0.972302 (rank : 5) | θ value | 2.24094e-75 (rank : 5) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P43694 | Gene names | GATA4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GATA4_MOUSE
|
||||||
| NC score | 0.970254 (rank : 6) | θ value | 8.51556e-75 (rank : 6) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q08369, P97491, Q9QZK4 | Gene names | Gata4, Gata-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
GATA1_MOUSE
|
||||||
| NC score | 0.948163 (rank : 7) | θ value | 1.23823e-49 (rank : 10) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17679 | Gene names | Gata1, Gf-1 | |||
|
Domain Architecture |

|
|||||
| Description | Erythroid transcription factor (GATA-binding factor 1) (GATA-1) (Eryf1) (GF-1) (NF-E1 DNA-binding protein). | |||||
|
GATA1_HUMAN
|
||||||
| NC score | 0.945371 (rank : 8) | θ value | 6.14528e-49 (rank : 12) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P15976, Q96GB8 | Gene names | GATA1, ERYF1, GF1 | |||
|
Domain Architecture |

|
|||||
| Description | Erythroid transcription factor (GATA-binding factor 1) (GATA-1) (Eryf1) (GF-1) (NF-E1 DNA-binding protein). | |||||
|
GATA3_MOUSE
|
||||||
| NC score | 0.932749 (rank : 9) | θ value | 1.32293e-51 (rank : 7) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P23772 | Gene names | Gata3, Gata-3 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-acting T-cell-specific transcription factor GATA-3 (GATA-binding factor 3). | |||||
|
GATA3_HUMAN
|
||||||
| NC score | 0.931318 (rank : 10) | θ value | 2.94719e-51 (rank : 8) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P23771, Q96J16 | Gene names | GATA3 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-acting T-cell-specific transcription factor GATA-3 (GATA-binding factor 3). | |||||
|
GATA2_HUMAN
|
||||||
| NC score | 0.924965 (rank : 11) | θ value | 3.2585e-50 (rank : 9) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P23769, Q9BUJ6 | Gene names | GATA2 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial transcription factor GATA-2 (GATA-binding protein 2). | |||||
|
GATA2_MOUSE
|
||||||
| NC score | 0.922369 (rank : 12) | θ value | 1.23823e-49 (rank : 11) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O09100 | Gene names | Gata2 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial transcription factor GATA-2 (GATA-binding protein 2). | |||||
|
TRPS1_HUMAN
|
||||||
| NC score | 0.478435 (rank : 13) | θ value | 1.2105e-12 (rank : 14) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UHF7, Q9NWE1, Q9UHH6 | Gene names | TRPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1 (Zinc finger protein GC79) (Tricho-rhino-phalangeal syndrome type I protein). | |||||
|
TRPS1_MOUSE
|
||||||
| NC score | 0.472515 (rank : 14) | θ value | 4.16044e-13 (rank : 13) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
CAN7_MOUSE
|
||||||
| NC score | 0.055007 (rank : 15) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R1S8, Q9Z0P9 | Gene names | Capn7, Palbh | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
HD_MOUSE
|
||||||
| NC score | 0.049329 (rank : 16) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
CAN7_HUMAN
|
||||||
| NC score | 0.048044 (rank : 17) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6W3 | Gene names | CAPN7, PALBH | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
HD_HUMAN
|
||||||
| NC score | 0.047747 (rank : 18) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.045258 (rank : 19) | θ value | 0.0961366 (rank : 15) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ZN533_HUMAN
|
||||||
| NC score | 0.045001 (rank : 20) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q569K4, Q49A04, Q6ZMZ7, Q8IY01, Q8N8H2, Q96DK4 | Gene names | ZNF533 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 533. | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.042553 (rank : 21) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
ZCH10_HUMAN
|
||||||
| NC score | 0.041572 (rank : 22) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.039465 (rank : 23) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
MTA2_HUMAN
|
||||||
| NC score | 0.038937 (rank : 24) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O94776, Q9UQB5 | Gene names | MTA2, MTA1L1, PID | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex). | |||||
|
MTA2_MOUSE
|
||||||
| NC score | 0.038932 (rank : 25) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R190, Q3TVT8 | Gene names | Mta2, Mta1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1). | |||||
|
ZN533_MOUSE
|
||||||
| NC score | 0.034840 (rank : 26) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BXJ8, Q8BWQ7 | Gene names | Znf533, Zfp533 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 533. | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.033832 (rank : 27) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
ZCH10_MOUSE
|
||||||
| NC score | 0.027322 (rank : 28) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.025811 (rank : 29) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
CDSN_HUMAN
|
||||||
| NC score | 0.024073 (rank : 30) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15517, O43509, Q5SQ85, Q5STD2, Q7LA70, Q7LA71, Q86Z04, Q8IZU4, Q8IZU5, Q8IZU6, Q8N5P3, Q95IF9, Q9NP52, Q9NPE0, Q9NPG5, Q9NRH4, Q9NRH5, Q9NRH6, Q9NRH7, Q9NRH8, Q9UBH8, Q9UIN6, Q9UIN7, Q9UIN8, Q9UIN9, Q9UIP0 | Gene names | CDSN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor (S protein). | |||||
|
MTSS1_HUMAN
|
||||||
| NC score | 0.023427 (rank : 31) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.021212 (rank : 32) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.020646 (rank : 33) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.019846 (rank : 34) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.019764 (rank : 35) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.018995 (rank : 36) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
TNC6B_HUMAN
|
||||||
| NC score | 0.017921 (rank : 37) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
MEF2B_MOUSE
|
||||||
| NC score | 0.017722 (rank : 38) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
RNPS1_HUMAN
|
||||||
| NC score | 0.017110 (rank : 39) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| NC score | 0.017110 (rank : 40) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.016898 (rank : 41) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.016802 (rank : 42) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.016775 (rank : 43) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.016134 (rank : 44) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
TSH3_HUMAN
|
||||||
| NC score | 0.015059 (rank : 45) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q63HK5, Q9H0G6, Q9P254 | Gene names | TSHZ3, KIAA1474, TSH3, ZNF537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.013551 (rank : 46) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.011737 (rank : 47) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.011173 (rank : 48) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
RPA2_MOUSE
|
||||||
| NC score | 0.010772 (rank : 49) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70700 | Gene names | Polr1b, Rpa2, Rpo1-2 | |||
|
Domain Architecture |
|
|||||
| Description | DNA-directed RNA polymerase I 135 kDa polypeptide (EC 2.7.7.6) (RNA polymerase I subunit 2) (RPA135). | |||||
|
NP1L3_MOUSE
|
||||||
| NC score | 0.008862 (rank : 50) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
MTMR5_HUMAN
|
||||||
| NC score | 0.008826 (rank : 51) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.008781 (rank : 52) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
OTX1_HUMAN
|
||||||
| NC score | 0.008631 (rank : 53) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P32242, Q53TG6 | Gene names | OTX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein OTX1. | |||||
|
CO4A2_HUMAN
|
||||||
| NC score | 0.008530 (rank : 54) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
AKAP2_HUMAN
|
||||||
| NC score | 0.007764 (rank : 55) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
NF2L1_HUMAN
|
||||||
| NC score | 0.007761 (rank : 56) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14494, Q12877, Q96FN6 | Gene names | NFE2L1, HBZ17, NRF1, TCF11 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1) (Transcription factor 11) (Transcription factor HBZ17) (Transcription factor LCR-F1) (Locus control region-factor 1). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.007586 (rank : 57) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
OTX1_MOUSE
|
||||||
| NC score | 0.007241 (rank : 58) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P80205 | Gene names | Otx1, Otx-1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein OTX1. | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.006906 (rank : 59) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
SOX14_HUMAN
|
||||||
| NC score | 0.006174 (rank : 60) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95416, Q3KPH7 | Gene names | SOX14, SOX28 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-14. | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.004418 (rank : 61) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
ZN11B_HUMAN
|
||||||
| NC score | 0.003776 (rank : 62) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 802 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q06732, Q86XY8, Q8NDW3 | Gene names | ZNF11B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 11B. | |||||
|
GBRG1_MOUSE
|
||||||
| NC score | 0.003341 (rank : 63) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0Y8 | Gene names | Gabrg1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric-acid receptor subunit gamma-1 precursor (GABA(A) receptor subunit gamma-1). | |||||
|
HOME1_HUMAN
|
||||||
| NC score | 0.002526 (rank : 64) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86YM7, O96003, Q86YM5 | Gene names | HOMER1, SYN47 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1. | |||||
|
TAXB1_HUMAN
|
||||||
| NC score | 0.002349 (rank : 65) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
ZN509_MOUSE
|
||||||
| NC score | 0.002079 (rank : 66) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
ZN672_MOUSE
|
||||||
| NC score | 0.001963 (rank : 67) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99LH4, Q8BIR7 | Gene names | Znf672, Zfp672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 672. | |||||
|
ZN306_HUMAN
|
||||||
| NC score | 0.001854 (rank : 68) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BRR0, Q5VXH3, Q92972, Q9H4T3 | Gene names | ZNF306, ZFP47, ZSCAN13 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 306 (Zinc finger protein 47 homolog) (Zfp-47) (Zf47) (Zinc finger and SCAN domain-containing protein 13). | |||||
|
ZN509_HUMAN
|
||||||
| NC score | 0.001740 (rank : 69) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ZSB9, Q59FJ4, Q5EBN0, Q8TB80 | Gene names | ZNF509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
ZN443_HUMAN
|
||||||
| NC score | 0.001702 (rank : 70) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2A4 | Gene names | ZNF443 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 443 (Krueppel-type zinc finger protein ZK1). | |||||
|
ZN442_HUMAN
|
||||||
| NC score | 0.001523 (rank : 71) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H7R0 | Gene names | ZNF442 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 442. | |||||
|
ERR2_MOUSE
|
||||||
| NC score | 0.001109 (rank : 72) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61539, O09067, O09146 | Gene names | Esrrb, Err-2, Err2, Nr3b2 | |||
|
Domain Architecture |

|
|||||
| Description | Steroid hormone receptor ERR2 (Estrogen-related receptor, beta) (ERR- beta) (Estrogen receptor-like 2). | |||||
|
ULK2_HUMAN
|
||||||
| NC score | -0.002184 (rank : 73) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 73 | Target Neighborhood Hits | 979 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYT8, O75119 | Gene names | ULK2, KIAA0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2). | |||||