Please be patient as the page loads
|
MYCN_MOUSE
|
||||||
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MYCN_MOUSE
|
||||||
| θ value | 1.41267e-170 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCN_HUMAN
|
||||||
| θ value | 4.72459e-150 (rank : 2) | NC score | 0.981433 (rank : 2) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCS_MOUSE
|
||||||
| θ value | 3.54468e-105 (rank : 3) | NC score | 0.955323 (rank : 3) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
MYC_MOUSE
|
||||||
| θ value | 3.06405e-32 (rank : 4) | NC score | 0.827098 (rank : 4) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYC_HUMAN
|
||||||
| θ value | 3.3877e-31 (rank : 5) | NC score | 0.820753 (rank : 5) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYCL1_HUMAN
|
||||||
| θ value | 2.27234e-27 (rank : 6) | NC score | 0.783242 (rank : 6) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL2_HUMAN
|
||||||
| θ value | 4.43474e-23 (rank : 7) | NC score | 0.745339 (rank : 7) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
MYCL1_MOUSE
|
||||||
| θ value | 5.07402e-19 (rank : 8) | NC score | 0.743898 (rank : 8) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCB_MOUSE
|
||||||
| θ value | 1.86753e-13 (rank : 9) | NC score | 0.694378 (rank : 9) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P8Z1, Q9D6E3 | Gene names | Mycb, Bmyc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein B-Myc. | |||||
|
MAX_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 10) | NC score | 0.617499 (rank : 10) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MAX_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 11) | NC score | 0.610014 (rank : 11) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
FIGLA_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 12) | NC score | 0.348104 (rank : 16) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O55208 | Gene names | Figla | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
SRBP2_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 13) | NC score | 0.289726 (rank : 24) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
MNT_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 14) | NC score | 0.303043 (rank : 21) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 15) | NC score | 0.310508 (rank : 18) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
FIGLA_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 16) | NC score | 0.331455 (rank : 17) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6QHK4 | Gene names | FIGLA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
ASCL1_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 17) | NC score | 0.269784 (rank : 28) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P50553, Q9BQ30 | Gene names | ASCL1, ASH1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (HASH1). | |||||
|
USF1_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 18) | NC score | 0.351928 (rank : 14) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 19) | NC score | 0.354385 (rank : 12) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
ASCL1_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 20) | NC score | 0.256316 (rank : 30) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q02067 | Gene names | Ascl1, Ash1, Mash-1, Mash1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (Mash-1). | |||||
|
MITF_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 21) | NC score | 0.276061 (rank : 26) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
USF2_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 22) | NC score | 0.352257 (rank : 13) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 23) | NC score | 0.349090 (rank : 15) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
MITF_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 24) | NC score | 0.273746 (rank : 27) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MAD1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 25) | NC score | 0.213767 (rank : 42) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MAD4_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 26) | NC score | 0.290986 (rank : 23) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
ASCL2_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 27) | NC score | 0.241067 (rank : 38) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35885, Q9WUJ7 | Gene names | Ascl2, Mash2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash-2). | |||||
|
PTF1A_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 28) | NC score | 0.196020 (rank : 46) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QX98, Q9QYF5, Q9QYF6 | Gene names | Ptf1a, Ptf1p48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
SRBP1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 29) | NC score | 0.253470 (rank : 34) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
BHLH8_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 30) | NC score | 0.246360 (rank : 36) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7RTS1 | Gene names | BHLHB8, MIST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
BHLH8_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 31) | NC score | 0.255069 (rank : 32) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
CBX2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 32) | NC score | 0.047900 (rank : 134) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
MXI1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 33) | NC score | 0.304409 (rank : 20) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
TFAP4_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 34) | NC score | 0.307312 (rank : 19) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 35) | NC score | 0.278323 (rank : 25) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
NDF2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 36) | NC score | 0.129453 (rank : 61) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15784, Q8TBI7, Q9UQC6 | Gene names | NEUROD2, NDRF | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
NDF2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 37) | NC score | 0.129310 (rank : 62) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62414, Q61952, Q925V5 | Gene names | Neurod2, Ndrf | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 38) | NC score | 0.238206 (rank : 39) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
ASCL2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 39) | NC score | 0.205623 (rank : 43) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99929, Q9UM68 | Gene names | ASCL2 | |||
|
Domain Architecture |
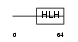
|
|||||
| Description | Achaete-scute homolog 2 (Mash2) (HASH2). | |||||
|
MAD4_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 40) | NC score | 0.241276 (rank : 37) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
ASCL3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 41) | NC score | 0.187848 (rank : 48) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NQ33, Q8WYQ6 | Gene names | ASCL3, HASH3, SGN1 | |||
|
Domain Architecture |
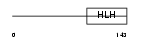
|
|||||
| Description | Achaete-scute homolog 3 (bHLH transcriptional regulator Sgn-1). | |||||
|
MLX_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 42) | NC score | 0.253691 (rank : 33) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 43) | NC score | 0.255636 (rank : 31) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MXI1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 44) | NC score | 0.291799 (rank : 22) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.163984 (rank : 45) | NC score | 0.026495 (rank : 143) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PTF1A_HUMAN
|
||||||
| θ value | 0.163984 (rank : 46) | NC score | 0.192966 (rank : 47) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7RTS3, Q9HC25 | Gene names | PTF1A, PTF1P48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 47) | NC score | 0.124522 (rank : 66) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 48) | NC score | 0.124979 (rank : 64) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 0.21417 (rank : 49) | NC score | 0.137061 (rank : 57) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 0.21417 (rank : 50) | NC score | 0.132797 (rank : 59) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ASCL3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 51) | NC score | 0.173938 (rank : 50) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JJR7 | Gene names | Ascl3, Mash3, Sgn1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 3 (bHLH transcriptional regulator Sgn-1) (Mash- 3). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 0.365318 (rank : 52) | NC score | 0.027703 (rank : 141) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
HAND2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 53) | NC score | 0.201774 (rank : 44) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P61296, O95300, O95301, P97833 | Gene names | HAND2, DHAND | |||
|
Domain Architecture |
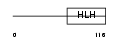
|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND). | |||||
|
HAND2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 54) | NC score | 0.201774 (rank : 45) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61039, Q61100 | Gene names | Hand2, Dhand, Hed, Thing2 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND) (Helix-loop-helix transcription factor expressed in embryo and deciduum) (Thing-2). | |||||
|
NGN3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 55) | NC score | 0.125341 (rank : 63) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y4Z2, Q9BY24 | Gene names | NEUROG3, NGN3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3. | |||||
|
OSBP2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 56) | NC score | 0.021909 (rank : 146) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q969R2, O60396, Q9BY96, Q9BZF0 | Gene names | OSBP2, KIAA1664, ORP4, OSBPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 2 (Oxysterol-binding protein-related protein 4) (OSBP-related protein 4) (ORP-4). | |||||
|
SDPR_MOUSE
|
||||||
| θ value | 0.47712 (rank : 57) | NC score | 0.033298 (rank : 138) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
TFEB_MOUSE
|
||||||
| θ value | 0.47712 (rank : 58) | NC score | 0.222917 (rank : 40) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
HAND1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 59) | NC score | 0.173198 (rank : 51) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O96004 | Gene names | HAND1, EHAND | |||
|
Domain Architecture |
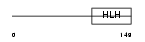
|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND). | |||||
|
HAND1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.178320 (rank : 49) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q64279, Q61099 | Gene names | Hand1, Ehand, Hxt, Thing1 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND) (Helix-loop-helix transcription factor expressed in extraembryonic mesoderm and trophoblast) (Thing-1) (Th1). | |||||
|
ITF2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 61) | NC score | 0.078589 (rank : 98) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P15884, Q15439, Q15440 | Gene names | TCF4, ITF2, SEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (SL3-3 enhancer factor 2) (SEF-2). | |||||
|
ITF2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 62) | NC score | 0.079760 (rank : 96) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q60722, Q60721, Q62211, Q64072, Q80UE8 | Gene names | Tcf4, Itf2, Sef2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (MITF-2) (SL3-3 enhancer factor 2) (SEF-2) (Class A helix-loop-helix transcription factor ME2). | |||||
|
TFEB_HUMAN
|
||||||
| θ value | 0.62314 (rank : 63) | NC score | 0.221242 (rank : 41) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
SPIB_MOUSE
|
||||||
| θ value | 0.813845 (rank : 64) | NC score | 0.032528 (rank : 139) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
HTF4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.071748 (rank : 107) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99081 | Gene names | TCF12, HEB, HTF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4). | |||||
|
FGD5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.015714 (rank : 161) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
HTF4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.070801 (rank : 110) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61286 | Gene names | Tcf12, Alf1, Meb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4) (Class A helix-loop-helix transcription factor ME1). | |||||
|
ID1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.120636 (rank : 68) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P41134, O00651, O00652, Q16371, Q16377, Q5TE66, Q5TE67, Q969Z7, Q9H0Z5, Q9H109 | Gene names | ID1, ID | |||
|
Domain Architecture |
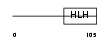
|
|||||
| Description | DNA-binding protein inhibitor ID-1 (Inhibitor of DNA binding 1). | |||||
|
ID1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.118358 (rank : 69) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P20067, Q61101, Q9D897 | Gene names | Id1, Id, Id-1, Idb1 | |||
|
Domain Architecture |
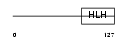
|
|||||
| Description | DNA-binding protein inhibitor ID-1 (Inhibitor of DNA binding 1). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.016695 (rank : 158) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
SMBP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.019593 (rank : 154) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
CSF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.023264 (rank : 144) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.012790 (rank : 173) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.019533 (rank : 155) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
DOCK6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.015477 (rank : 165) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96HP0, Q9P2F2 | Gene names | DOCK6, KIAA1395 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 6. | |||||
|
GATA6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.012902 (rank : 172) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92908, P78327 | Gene names | GATA6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
GDIA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.040669 (rank : 135) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31150, P50394, Q7Z2G6, Q7Z2G9, Q7Z2H5, Q7Z2I6 | Gene names | GDI1, OPHN2, RABGDIA, XAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rab GDP dissociation inhibitor alpha (Rab GDI alpha) (Guanosine diphosphate dissociation inhibitor 1) (GDI-1) (XAP-4) (Oligophrenin- 2). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.012193 (rank : 179) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.020880 (rank : 153) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.003311 (rank : 188) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
MAD3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.257458 (rank : 29) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MAD3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.250521 (rank : 35) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.014294 (rank : 170) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.012709 (rank : 175) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
PODXL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.027458 (rank : 142) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
RL7_MOUSE
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.021425 (rank : 151) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P14148, Q80UR0 | Gene names | Rpl7 | |||
|
Domain Architecture |

|
|||||
| Description | 60S ribosomal protein L7. | |||||
|
ATOH1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.124728 (rank : 65) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92858 | Gene names | ATOH1, ATH1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein hATH-1). | |||||
|
ATOH1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.124503 (rank : 67) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P48985 | Gene names | Atoh1, Ath1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein mATH-1) (MATH1). | |||||
|
EP300_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.012712 (rank : 174) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
GDIA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.037586 (rank : 136) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P50396, Q8VHM3, Q91Y71, Q96CX5 | Gene names | Gdi1, Rabgdia | |||
|
Domain Architecture |
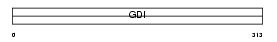
|
|||||
| Description | Rab GDP dissociation inhibitor alpha (Rab GDI alpha) (Guanosine diphosphate dissociation inhibitor 1) (GDI-1). | |||||
|
GDIB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.037273 (rank : 137) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61598, Q541Z9, Q8C530, Q9D8M9 | Gene names | Gdi2, Gdi3 | |||
|
Domain Architecture |
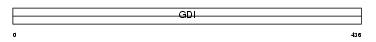
|
|||||
| Description | Rab GDP dissociation inhibitor beta (Rab GDI beta) (Guanosine diphosphate dissociation inhibitor 2) (GDI-2) (GDI-3). | |||||
|
SMC4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.014371 (rank : 169) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
WNK4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.000651 (rank : 190) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
CASP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.015318 (rank : 166) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.008525 (rank : 183) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.016564 (rank : 159) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
GAK11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.021588 (rank : 149) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.022012 (rank : 145) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.021519 (rank : 150) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.021595 (rank : 148) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.021726 (rank : 147) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
RHG05_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.015825 (rank : 160) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RHG05_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.015490 (rank : 163) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
SRCA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.012508 (rank : 176) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TEF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.017310 (rank : 157) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q10587, Q15729, Q7Z3J7, Q8IU94, Q96TG4 | Gene names | TEF | |||
|
Domain Architecture |

|
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
TEF_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.017371 (rank : 156) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLC6, Q3U426, Q3UGF4, Q3UM49, Q3URC8, Q6QHT6, Q8C6I0, Q8VD02 | Gene names | Tef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
TTLL4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.011904 (rank : 180) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14679, Q8WW29 | Gene names | TTLL4, KIAA0173 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
BHLH2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.058043 (rank : 121) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
BHLH2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.058932 (rank : 119) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
CASP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.015042 (rank : 168) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CEBPB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.015256 (rank : 167) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17676, Q96IH2, Q9H4Z5 | Gene names | CEBPB, TCF5 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Nuclear factor NF- IL6) (Transcription factor 5). | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.015480 (rank : 164) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
GAK10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.021065 (rank : 152) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.029409 (rank : 140) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
ALS2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.006039 (rank : 184) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
ARID2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.013005 (rank : 171) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
EPN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.015490 (rank : 162) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
ETV3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.009200 (rank : 182) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
KCNC4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.002199 (rank : 189) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KI21A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.005884 (rank : 185) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.005660 (rank : 186) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
PEPL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.010361 (rank : 181) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
SNPC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.012224 (rank : 178) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91XA5, Q3UK17, Q8CBS9 | Gene names | Snapc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 2 (SNAPc subunit 2) (snRNA- activating protein complex 45 kDa subunit) (SNAPc 45 kDa subunit) (Small nuclear RNA-activating complex polypeptide 2). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.012505 (rank : 177) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
UN45B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.005548 (rank : 187) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CGY6, Q5XG72, Q8BHC5, Q8BWK3 | Gene names | Unc45b, Cmya4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
BHLH3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.060665 (rank : 115) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
BHLH3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.056521 (rank : 123) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
BHLH4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.060230 (rank : 116) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NDY6 | Gene names | BHLHB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 4 (bHLHB4). | |||||
|
BHLH4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.058934 (rank : 118) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BGW3, Q71MB7 | Gene names | Bhlhb4, H20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 4 (bHLHB4). | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.077291 (rank : 100) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.076843 (rank : 101) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
HEN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.096454 (rank : 79) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02575 | Gene names | NHLH1, HEN1 | |||
|
Domain Architecture |
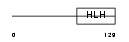
|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
HEN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.098501 (rank : 76) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02576 | Gene names | Nhlh1, Hen1 | |||
|
Domain Architecture |
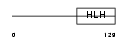
|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
HEN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.096150 (rank : 80) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02577 | Gene names | NHLH2, HEN2 | |||
|
Domain Architecture |
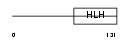
|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
HEN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.095343 (rank : 81) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q64221 | Gene names | Nhlh2, Hen2 | |||
|
Domain Architecture |
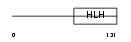
|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
HEY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.051248 (rank : 132) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.052266 (rank : 130) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.054905 (rank : 127) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.055286 (rank : 125) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
ID2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.056832 (rank : 122) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02363 | Gene names | ID2 | |||
|
Domain Architecture |
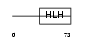
|
|||||
| Description | DNA-binding protein inhibitor ID-2 (Inhibitor of DNA binding 2). | |||||
|
ID2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.051398 (rank : 131) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P41136, O88604 | Gene names | Id2, Id-2, Idb2 | |||
|
Domain Architecture |
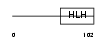
|
|||||
| Description | DNA-binding protein inhibitor ID-2 (Inhibitor of DNA binding 2). | |||||
|
ID4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.067031 (rank : 112) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P47928, Q13005 | Gene names | ID4 | |||
|
Domain Architecture |
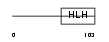
|
|||||
| Description | DNA-binding protein inhibitor ID-4 (Inhibitor of DNA binding 4). | |||||
|
ID4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.065074 (rank : 113) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P41139 | Gene names | Id4, Id-4, Idb4 | |||
|
Domain Architecture |
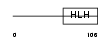
|
|||||
| Description | DNA-binding protein inhibitor ID-4 (Inhibitor of DNA binding 4). | |||||
|
LYL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.098179 (rank : 78) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P12980, O76102 | Gene names | LYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
LYL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.099619 (rank : 75) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P27792 | Gene names | Lyl1, Lyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
MAD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.148387 (rank : 52) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MUSC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.076416 (rank : 103) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60682, O75946, Q9BRE7 | Gene names | MSC, ABF1 | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Activated B-cell factor 1) (ABF-1). | |||||
|
MUSC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.077565 (rank : 99) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O88940 | Gene names | Msc, Myor | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Myogenic repressor). | |||||
|
MYF5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.080908 (rank : 94) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P13349 | Gene names | MYF5 | |||
|
Domain Architecture |
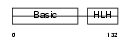
|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
MYF5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.082805 (rank : 92) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P24699, Q543W7 | Gene names | Myf5, Myf-5 | |||
|
Domain Architecture |
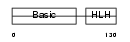
|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
MYF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.093267 (rank : 83) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P23409, Q53X80, Q6FHI9 | Gene names | MYF6 | |||
|
Domain Architecture |
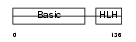
|
|||||
| Description | Myogenic factor 6 (Myf-6). | |||||
|
MYF6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.089169 (rank : 86) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P15375 | Gene names | Myf6, Myf-6 | |||
|
Domain Architecture |
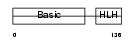
|
|||||
| Description | Myogenic factor 6 (Myf-6) (Herculin). | |||||
|
MYOD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.076754 (rank : 102) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P15172 | Gene names | MYOD1, MYF3, MYOD | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1 (Myogenic factor 3) (Myf-3). | |||||
|
MYOD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.079363 (rank : 97) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P10085 | Gene names | Myod1, Myod | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1. | |||||
|
MYOG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.088485 (rank : 88) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P15173 | Gene names | MYOG, MYF4 | |||
|
Domain Architecture |
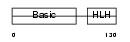
|
|||||
| Description | Myogenin (Myogenic factor 4) (Myf-4). | |||||
|
MYOG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.089004 (rank : 87) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P12979 | Gene names | Myog | |||
|
Domain Architecture |
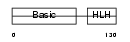
|
|||||
| Description | Myogenin (MYOD1-related protein). | |||||
|
NDF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.085768 (rank : 90) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
NDF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.086968 (rank : 89) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
NDF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.070677 (rank : 111) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9HD90 | Gene names | NEUROD4 | |||
|
Domain Architecture |
|
|||||
| Description | Neurogenic differentiation factor 4 (NeuroD4). | |||||
|
NDF4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.071490 (rank : 109) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O09105 | Gene names | Neurod4, Ath3, Atoh3 | |||
|
Domain Architecture |
|
|||||
| Description | Neurogenic differentiation factor 4 (NeuroD4) (Atonal protein homolog 3) (Helix-loop-helix protein mATH-3) (MATH3). | |||||
|
NDF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.072073 (rank : 105) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96NK8, Q9H3H6 | Gene names | NEUROD6 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 6 (NeuroD6). | |||||
|
NDF6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.071980 (rank : 106) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P48986 | Gene names | Neurod6, Ath2, Atoh2, Nex1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 6 (NeuroD6) (Atonal protein homolog 2) (Helix-loop-helix protein mATH-2) (MATH2) (NEX-1 protein). | |||||
|
NGN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.080574 (rank : 95) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92886, Q96HE1 | Gene names | NEUROG1, NEUROD3, NGN, NGN1 | |||
|
Domain Architecture |
|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein). | |||||
|
NGN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.082063 (rank : 93) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70660 | Gene names | Neurog1, Ath4c, Neurod3, Ngn, Ngn1 | |||
|
Domain Architecture |
|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein) (Helix-loop-helix protein mATH-4C) (MATH4C). | |||||
|
NGN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.100614 (rank : 74) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H2A3, Q8N416 | Gene names | NEUROG2, NGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogenin 2. | |||||
|
NGN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.100723 (rank : 73) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70447, P70237 | Gene names | Neurog2, Ath4a, Atoh4, Ngn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 2 (Atonal protein homolog 4) (Helix-loop-helix protein mATH-4A) (MATH4A). | |||||
|
NGN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.101833 (rank : 72) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70661 | Gene names | Neurog3, Ath4b, Atoh5, Ngn3 | |||
|
Domain Architecture |
|
|||||
| Description | Neurogenin 3 (Atonal protein homolog 5) (Helix-loop-helix protein mATH-4B) (MATH4B). | |||||
|
OLIG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.054883 (rank : 128) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TAK6, Q7RTS0 | Gene names | OLIG1 | |||
|
Domain Architecture |
|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1). | |||||
|
OLIG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.054573 (rank : 129) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JKN5 | Gene names | Olig1 | |||
|
Domain Architecture |
|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1) (Oligodendrocyte- specific bHLH transcription factor 1). | |||||
|
OLIG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.058995 (rank : 117) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13516, Q86X04, Q9NZ14 | Gene names | OLIG2, BHLHB1, PRKCBP2, RACK17 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2) (Basic helix-loop- helix protein class B 1) (Protein kinase C-binding protein RACK17) (Protein kinase C-binding protein 2). | |||||
|
OLIG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.058208 (rank : 120) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9EQW6, Q9JKN4 | Gene names | Olig2 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2). | |||||
|
OLIG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.055038 (rank : 126) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7RTU3, Q8N8Q0 | Gene names | OLIG3, BHLHB7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oligodendrocyte transcription factor 3 (Oligo3) (Basic helix-loop- helix domain-containing class B protein 7). | |||||
|
OLIG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.055870 (rank : 124) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PFG8, Q9EQW5 | Gene names | Olig3, Bhlhb7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oligodendrocyte transcription factor 3 (Oligo3) (Oligodendrocyte- specific bHLH transcription factor 3) (Basic helix-loop-helix domain- containing class B protein 7). | |||||
|
SCX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.132803 (rank : 58) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q64124 | Gene names | Scx | |||
|
Domain Architecture |
|
|||||
| Description | Basic helix-loop-helix transcription factor scleraxis. | |||||
|
TAL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.106953 (rank : 71) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P17542 | Gene names | TAL1, SCL, TCL5 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein (TAL-1 protein) (Stem cell protein) (T-cell leukemia/lymphoma-5 protein). | |||||
|
TAL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.108605 (rank : 70) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P22091 | Gene names | Tal1, Scl, Tal-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein homolog (TAL-1 protein) (Stem cell protein). | |||||
|
TAL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.092043 (rank : 84) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q16559 | Gene names | TAL2 | |||
|
Domain Architecture |
|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein (TAL-2 protein). | |||||
|
TAL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.091326 (rank : 85) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62282 | Gene names | Tal2 | |||
|
Domain Architecture |
|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein homolog (TAL-2 protein). | |||||
|
TCF15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.094324 (rank : 82) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q12870, Q9NQQ1 | Gene names | TCF15, BHLHEC2 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis). | |||||
|
TCF15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.098358 (rank : 77) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q60756, Q60788 | Gene names | Tcf15, Bhlhec2 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis) (Meso1). | |||||
|
TCF21_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.072496 (rank : 104) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43680, O43545, Q9BZ14 | Gene names | TCF21, POD1 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor 21 (Podocyte-expressed 1) (Pod-1) (Epicardin) (Capsulin). | |||||
|
TCF21_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.071623 (rank : 108) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35437, Q3U023 | Gene names | Tcf21, Pod1 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor 21 (Podocyte-expressed 1) (Pod-1) (Epicardin) (Capsulin). | |||||
|
TCFL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.062015 (rank : 114) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
TFE2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.050357 (rank : 133) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P15923, P15883, Q14208, Q14635, Q14636, Q9UPI9 | Gene names | TCF3, E2A, ITF1 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Immunoglobulin transcription factor 1) (Transcription factor ITF-1) (Kappa-E2-binding factor). | |||||
|
TWST1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.141529 (rank : 55) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15672, Q92487, Q99804 | Gene names | TWIST1, TWIST | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (H-twist). | |||||
|
TWST1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.141388 (rank : 56) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P26687 | Gene names | Twist1, Twist | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (M-twist). | |||||
|
TWST2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.142476 (rank : 53) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WVJ9 | Gene names | TWIST2, DERMO1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.142469 (rank : 54) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D030 | Gene names | Twist2, Dermo1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
WBS14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.132359 (rank : 60) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
WBS14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.083322 (rank : 91) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
MYCN_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.41267e-170 (rank : 1) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCN_HUMAN
|
||||||
| NC score | 0.981433 (rank : 2) | θ value | 4.72459e-150 (rank : 2) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCS_MOUSE
|
||||||
| NC score | 0.955323 (rank : 3) | θ value | 3.54468e-105 (rank : 3) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
MYC_MOUSE
|
||||||
| NC score | 0.827098 (rank : 4) | θ value | 3.06405e-32 (rank : 4) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYC_HUMAN
|
||||||
| NC score | 0.820753 (rank : 5) | θ value | 3.3877e-31 (rank : 5) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYCL1_HUMAN
|
||||||
| NC score | 0.783242 (rank : 6) | θ value | 2.27234e-27 (rank : 6) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL2_HUMAN
|
||||||
| NC score | 0.745339 (rank : 7) | θ value | 4.43474e-23 (rank : 7) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
MYCL1_MOUSE
|
||||||
| NC score | 0.743898 (rank : 8) | θ value | 5.07402e-19 (rank : 8) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCB_MOUSE
|
||||||
| NC score | 0.694378 (rank : 9) | θ value | 1.86753e-13 (rank : 9) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P8Z1, Q9D6E3 | Gene names | Mycb, Bmyc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein B-Myc. | |||||
|
MAX_HUMAN
|
||||||
| NC score | 0.617499 (rank : 10) | θ value | 3.64472e-09 (rank : 10) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MAX_MOUSE
|
||||||
| NC score | 0.610014 (rank : 11) | θ value | 1.06045e-08 (rank : 11) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.354385 (rank : 12) | θ value | 0.00102713 (rank : 19) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.352257 (rank : 13) | θ value | 0.00228821 (rank : 22) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.351928 (rank : 14) | θ value | 0.00102713 (rank : 18) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_MOUSE
|
||||||
| NC score | 0.349090 (rank : 15) | θ value | 0.00228821 (rank : 23) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
FIGLA_MOUSE
|
||||||
| NC score | 0.348104 (rank : 16) | θ value | 4.92598e-06 (rank : 12) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O55208 | Gene names | Figla | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
FIGLA_HUMAN
|
||||||
| NC score | 0.331455 (rank : 17) | θ value | 0.00035302 (rank : 16) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6QHK4 | Gene names | FIGLA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.310508 (rank : 18) | θ value | 0.00020696 (rank : 15) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
TFAP4_HUMAN
|
||||||
| NC score | 0.307312 (rank : 19) | θ value | 0.0431538 (rank : 34) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
MXI1_MOUSE
|
||||||
| NC score | 0.304409 (rank : 20) | θ value | 0.0431538 (rank : 33) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.303043 (rank : 21) | θ value | 0.000158464 (rank : 14) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MXI1_HUMAN
|
||||||
| NC score | 0.291799 (rank : 22) | θ value | 0.163984 (rank : 44) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MAD4_HUMAN
|
||||||
| NC score | 0.290986 (rank : 23) | θ value | 0.0113563 (rank : 26) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
SRBP2_HUMAN
|
||||||
| NC score | 0.289726 (rank : 24) | θ value | 9.29e-05 (rank : 13) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.278323 (rank : 25) | θ value | 0.0431538 (rank : 35) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.276061 (rank : 26) | θ value | 0.00228821 (rank : 21) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.273746 (rank : 27) | θ value | 0.00390308 (rank : 24) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
ASCL1_HUMAN
|
||||||
| NC score | 0.269784 (rank : 28) | θ value | 0.00102713 (rank : 17) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P50553, Q9BQ30 | Gene names | ASCL1, ASH1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (HASH1). | |||||
|
MAD3_HUMAN
|
||||||
| NC score | 0.257458 (rank : 29) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
ASCL1_MOUSE
|
||||||
| NC score | 0.256316 (rank : 30) | θ value | 0.00228821 (rank : 20) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q02067 | Gene names | Ascl1, Ash1, Mash-1, Mash1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (Mash-1). | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.255636 (rank : 31) | θ value | 0.0961366 (rank : 43) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
BHLH8_MOUSE
|
||||||
| NC score | 0.255069 (rank : 32) | θ value | 0.0252991 (rank : 31) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.253691 (rank : 33) | θ value | 0.0961366 (rank : 42) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
SRBP1_HUMAN
|
||||||
| NC score | 0.253470 (rank : 34) | θ value | 0.0193708 (rank : 29) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
MAD3_MOUSE
|
||||||
| NC score | 0.250521 (rank : 35) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
BHLH8_HUMAN
|
||||||
| NC score | 0.246360 (rank : 36) | θ value | 0.0252991 (rank : 30) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7RTS1 | Gene names | BHLHB8, MIST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
MAD4_MOUSE
|
||||||
| NC score | 0.241276 (rank : 37) | θ value | 0.0736092 (rank : 40) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
ASCL2_MOUSE
|
||||||
| NC score | 0.241067 (rank : 38) | θ value | 0.0193708 (rank : 27) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35885, Q9WUJ7 | Gene names | Ascl2, Mash2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash-2). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.238206 (rank : 39) | θ value | 0.0563607 (rank : 38) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.222917 (rank : 40) | θ value | 0.47712 (rank : 58) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.221242 (rank : 41) | θ value | 0.62314 (rank : 63) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
MAD1_MOUSE
|
||||||
| NC score | 0.213767 (rank : 42) | θ value | 0.00869519 (rank : 25) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
ASCL2_HUMAN
|
||||||
| NC score | 0.205623 (rank : 43) | θ value | 0.0736092 (rank : 39) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99929, Q9UM68 | Gene names | ASCL2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash2) (HASH2). | |||||
|
HAND2_HUMAN
|
||||||
| NC score | 0.201774 (rank : 44) | θ value | 0.47712 (rank : 53) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P61296, O95300, O95301, P97833 | Gene names | HAND2, DHAND | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND). | |||||
|
HAND2_MOUSE
|
||||||
| NC score | 0.201774 (rank : 45) | θ value | 0.47712 (rank : 54) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61039, Q61100 | Gene names | Hand2, Dhand, Hed, Thing2 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND) (Helix-loop-helix transcription factor expressed in embryo and deciduum) (Thing-2). | |||||
|
PTF1A_MOUSE
|
||||||
| NC score | 0.196020 (rank : 46) | θ value | 0.0193708 (rank : 28) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QX98, Q9QYF5, Q9QYF6 | Gene names | Ptf1a, Ptf1p48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
PTF1A_HUMAN
|
||||||
| NC score | 0.192966 (rank : 47) | θ value | 0.163984 (rank : 46) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7RTS3, Q9HC25 | Gene names | PTF1A, PTF1P48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
ASCL3_HUMAN
|
||||||
| NC score | 0.187848 (rank : 48) | θ value | 0.0961366 (rank : 41) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NQ33, Q8WYQ6 | Gene names | ASCL3, HASH3, SGN1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 3 (bHLH transcriptional regulator Sgn-1). | |||||
|
HAND1_MOUSE
|
||||||
| NC score | 0.178320 (rank : 49) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q64279, Q61099 | Gene names | Hand1, Ehand, Hxt, Thing1 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND) (Helix-loop-helix transcription factor expressed in extraembryonic mesoderm and trophoblast) (Thing-1) (Th1). | |||||
|
ASCL3_MOUSE
|
||||||
| NC score | 0.173938 (rank : 50) | θ value | 0.279714 (rank : 51) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JJR7 | Gene names | Ascl3, Mash3, Sgn1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 3 (bHLH transcriptional regulator Sgn-1) (Mash- 3). | |||||
|
HAND1_HUMAN
|
||||||
| NC score | 0.173198 (rank : 51) | θ value | 0.62314 (rank : 59) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O96004 | Gene names | HAND1, EHAND | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND). | |||||
|
MAD1_HUMAN
|
||||||
| NC score | 0.148387 (rank : 52) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
TWST2_HUMAN
|
||||||
| NC score | 0.142476 (rank : 53) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WVJ9 | Gene names | TWIST2, DERMO1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST2_MOUSE
|
||||||
| NC score | 0.142469 (rank : 54) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D030 | Gene names | Twist2, Dermo1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST1_HUMAN
|
||||||
| NC score | 0.141529 (rank : 55) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15672, Q92487, Q99804 | Gene names | TWIST1, TWIST | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (H-twist). | |||||
|
TWST1_MOUSE
|
||||||
| NC score | 0.141388 (rank : 56) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P26687 | Gene names | Twist1, Twist | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (M-twist). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.137061 (rank : 57) | θ value | 0.21417 (rank : 49) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
SCX_MOUSE
|
||||||
| NC score | 0.132803 (rank : 58) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q64124 | Gene names | Scx | |||
|
Domain Architecture |

|
|||||
| Description | Basic helix-loop-helix transcription factor scleraxis. | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.132797 (rank : 59) | θ value | 0.21417 (rank : 50) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
WBS14_HUMAN
|
||||||
| NC score | 0.132359 (rank : 60) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
NDF2_HUMAN
|
||||||
| NC score | 0.129453 (rank : 61) | θ value | 0.0563607 (rank : 36) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15784, Q8TBI7, Q9UQC6 | Gene names | NEUROD2, NDRF | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
NDF2_MOUSE
|
||||||
| NC score | 0.129310 (rank : 62) | θ value | 0.0563607 (rank : 37) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62414, Q61952, Q925V5 | Gene names | Neurod2, Ndrf | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
NGN3_HUMAN
|
||||||
| NC score | 0.125341 (rank : 63) | θ value | 0.47712 (rank : 55) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y4Z2, Q9BY24 | Gene names | NEUROG3, NGN3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3. | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.124979 (rank : 64) | θ value | 0.21417 (rank : 48) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ATOH1_HUMAN
|
||||||
| NC score | 0.124728 (rank : 65) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92858 | Gene names | ATOH1, ATH1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein hATH-1). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.124522 (rank : 66) | θ value | 0.21417 (rank : 47) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ATOH1_MOUSE
|
||||||
| NC score | 0.124503 (rank : 67) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P48985 | Gene names | Atoh1, Ath1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein mATH-1) (MATH1). | |||||
|
ID1_HUMAN
|
||||||
| NC score | 0.120636 (rank : 68) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P41134, O00651, O00652, Q16371, Q16377, Q5TE66, Q5TE67, Q969Z7, Q9H0Z5, Q9H109 | Gene names | ID1, ID | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein inhibitor ID-1 (Inhibitor of DNA binding 1). | |||||
|
ID1_MOUSE
|
||||||
| NC score | 0.118358 (rank : 69) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P20067, Q61101, Q9D897 | Gene names | Id1, Id, Id-1, Idb1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein inhibitor ID-1 (Inhibitor of DNA binding 1). | |||||
|
TAL1_MOUSE
|
||||||
| NC score | 0.108605 (rank : 70) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P22091 | Gene names | Tal1, Scl, Tal-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein homolog (TAL-1 protein) (Stem cell protein). | |||||
|
TAL1_HUMAN
|
||||||
| NC score | 0.106953 (rank : 71) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P17542 | Gene names | TAL1, SCL, TCL5 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein (TAL-1 protein) (Stem cell protein) (T-cell leukemia/lymphoma-5 protein). | |||||
|
NGN3_MOUSE
|
||||||
| NC score | 0.101833 (rank : 72) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70661 | Gene names | Neurog3, Ath4b, Atoh5, Ngn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3 (Atonal protein homolog 5) (Helix-loop-helix protein mATH-4B) (MATH4B). | |||||
|
NGN2_MOUSE
|
||||||
| NC score | 0.100723 (rank : 73) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70447, P70237 | Gene names | Neurog2, Ath4a, Atoh4, Ngn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 2 (Atonal protein homolog 4) (Helix-loop-helix protein mATH-4A) (MATH4A). | |||||
|
NGN2_HUMAN
|
||||||
| NC score | 0.100614 (rank : 74) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H2A3, Q8N416 | Gene names | NEUROG2, NGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogenin 2. | |||||
|
LYL1_MOUSE
|
||||||
| NC score | 0.099619 (rank : 75) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P27792 | Gene names | Lyl1, Lyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
HEN1_MOUSE
|
||||||
| NC score | 0.098501 (rank : 76) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02576 | Gene names | Nhlh1, Hen1 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
TCF15_MOUSE
|
||||||
| NC score | 0.098358 (rank : 77) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q60756, Q60788 | Gene names | Tcf15, Bhlhec2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis) (Meso1). | |||||
|
LYL1_HUMAN
|
||||||
| NC score | 0.098179 (rank : 78) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P12980, O76102 | Gene names | LYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
HEN1_HUMAN
|
||||||
| NC score | 0.096454 (rank : 79) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02575 | Gene names | NHLH1, HEN1 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
HEN2_HUMAN
|
||||||
| NC score | 0.096150 (rank : 80) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02577 | Gene names | NHLH2, HEN2 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
HEN2_MOUSE
|
||||||
| NC score | 0.095343 (rank : 81) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q64221 | Gene names | Nhlh2, Hen2 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
TCF15_HUMAN
|
||||||
| NC score | 0.094324 (rank : 82) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q12870, Q9NQQ1 | Gene names | TCF15, BHLHEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis). | |||||
|
MYF6_HUMAN
|
||||||
| NC score | 0.093267 (rank : 83) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P23409, Q53X80, Q6FHI9 | Gene names | MYF6 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 6 (Myf-6). | |||||
|
TAL2_HUMAN
|
||||||
| NC score | 0.092043 (rank : 84) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q16559 | Gene names | TAL2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein (TAL-2 protein). | |||||
|
TAL2_MOUSE
|
||||||
| NC score | 0.091326 (rank : 85) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62282 | Gene names | Tal2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein homolog (TAL-2 protein). | |||||
|
MYF6_MOUSE
|
||||||
| NC score | 0.089169 (rank : 86) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P15375 | Gene names | Myf6, Myf-6 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 6 (Myf-6) (Herculin). | |||||
|
MYOG_MOUSE
|
||||||
| NC score | 0.089004 (rank : 87) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P12979 | Gene names | Myog | |||
|
Domain Architecture |

|
|||||
| Description | Myogenin (MYOD1-related protein). | |||||
|
MYOG_HUMAN
|
||||||
| NC score | 0.088485 (rank : 88) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P15173 | Gene names | MYOG, MYF4 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenin (Myogenic factor 4) (Myf-4). | |||||
|
NDF1_MOUSE
|
||||||
| NC score | 0.086968 (rank : 89) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
NDF1_HUMAN
|
||||||
| NC score | 0.085768 (rank : 90) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
WBS14_MOUSE
|
||||||
| NC score | 0.083322 (rank : 91) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
MYF5_MOUSE
|
||||||
| NC score | 0.082805 (rank : 92) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P24699, Q543W7 | Gene names | Myf5, Myf-5 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
NGN1_MOUSE
|
||||||
| NC score | 0.082063 (rank : 93) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70660 | Gene names | Neurog1, Ath4c, Neurod3, Ngn, Ngn1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein) (Helix-loop-helix protein mATH-4C) (MATH4C). | |||||
|
MYF5_HUMAN
|
||||||
| NC score | 0.080908 (rank : 94) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P13349 | Gene names | MYF5 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
NGN1_HUMAN
|
||||||
| NC score | 0.080574 (rank : 95) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92886, Q96HE1 | Gene names | NEUROG1, NEUROD3, NGN, NGN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein). | |||||
|
ITF2_MOUSE
|
||||||
| NC score | 0.079760 (rank : 96) | θ value | 0.62314 (rank : 62) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q60722, Q60721, Q62211, Q64072, Q80UE8 | Gene names | Tcf4, Itf2, Sef2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (MITF-2) (SL3-3 enhancer factor 2) (SEF-2) (Class A helix-loop-helix transcription factor ME2). | |||||
|
MYOD1_MOUSE
|
||||||
| NC score | 0.079363 (rank : 97) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P10085 | Gene names | Myod1, Myod | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1. | |||||
|
ITF2_HUMAN
|
||||||
| NC score | 0.078589 (rank : 98) | θ value | 0.62314 (rank : 61) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P15884, Q15439, Q15440 | Gene names | TCF4, ITF2, SEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (SL3-3 enhancer factor 2) (SEF-2). | |||||
|
MUSC_MOUSE
|
||||||
| NC score | 0.077565 (rank : 99) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O88940 | Gene names | Msc, Myor | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Myogenic repressor). | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.077291 (rank : 100) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.076843 (rank : 101) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
MYOD1_HUMAN
|
||||||
| NC score | 0.076754 (rank : 102) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P15172 | Gene names | MYOD1, MYF3, MYOD | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1 (Myogenic factor 3) (Myf-3). | |||||
|
MUSC_HUMAN
|
||||||
| NC score | 0.076416 (rank : 103) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60682, O75946, Q9BRE7 | Gene names | MSC, ABF1 | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Activated B-cell factor 1) (ABF-1). | |||||
|
TCF21_HUMAN
|
||||||
| NC score | 0.072496 (rank : 104) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43680, O43545, Q9BZ14 | Gene names | TCF21, POD1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 21 (Podocyte-expressed 1) (Pod-1) (Epicardin) (Capsulin). | |||||
|
NDF6_HUMAN
|
||||||
| NC score | 0.072073 (rank : 105) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96NK8, Q9H3H6 | Gene names | NEUROD6 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 6 (NeuroD6). | |||||
|
NDF6_MOUSE
|
||||||
| NC score | 0.071980 (rank : 106) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P48986 | Gene names | Neurod6, Ath2, Atoh2, Nex1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 6 (NeuroD6) (Atonal protein homolog 2) (Helix-loop-helix protein mATH-2) (MATH2) (NEX-1 protein). | |||||
|
HTF4_HUMAN
|
||||||
| NC score | 0.071748 (rank : 107) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99081 | Gene names | TCF12, HEB, HTF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4). | |||||
|
TCF21_MOUSE
|
||||||
| NC score | 0.071623 (rank : 108) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35437, Q3U023 | Gene names | Tcf21, Pod1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 21 (Podocyte-expressed 1) (Pod-1) (Epicardin) (Capsulin). | |||||
|
NDF4_MOUSE
|
||||||
| NC score | 0.071490 (rank : 109) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O09105 | Gene names | Neurod4, Ath3, Atoh3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 4 (NeuroD4) (Atonal protein homolog 3) (Helix-loop-helix protein mATH-3) (MATH3). | |||||
|
HTF4_MOUSE
|
||||||
| NC score | 0.070801 (rank : 110) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61286 | Gene names | Tcf12, Alf1, Meb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4) (Class A helix-loop-helix transcription factor ME1). | |||||
|
NDF4_HUMAN
|
||||||
| NC score | 0.070677 (rank : 111) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9HD90 | Gene names | NEUROD4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 4 (NeuroD4). | |||||
|
ID4_HUMAN
|
||||||
| NC score | 0.067031 (rank : 112) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P47928, Q13005 | Gene names | ID4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein inhibitor ID-4 (Inhibitor of DNA binding 4). | |||||
|
ID4_MOUSE
|
||||||
| NC score | 0.065074 (rank : 113) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P41139 | Gene names | Id4, Id-4, Idb4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein inhibitor ID-4 (Inhibitor of DNA binding 4). | |||||
|
TCFL5_HUMAN
|
||||||
| NC score | 0.062015 (rank : 114) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
BHLH3_HUMAN
|
||||||
| NC score | 0.060665 (rank : 115) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
BHLH4_HUMAN
|
||||||
| NC score | 0.060230 (rank : 116) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NDY6 | Gene names | BHLHB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 4 (bHLHB4). | |||||
|
OLIG2_HUMAN
|
||||||
| NC score | 0.058995 (rank : 117) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13516, Q86X04, Q9NZ14 | Gene names | OLIG2, BHLHB1, PRKCBP2, RACK17 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2) (Basic helix-loop- helix protein class B 1) (Protein kinase C-binding protein RACK17) (Protein kinase C-binding protein 2). | |||||
|
BHLH4_MOUSE
|
||||||
| NC score | 0.058934 (rank : 118) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BGW3, Q71MB7 | Gene names | Bhlhb4, H20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 4 (bHLHB4). | |||||
|
BHLH2_MOUSE
|
||||||
| NC score | 0.058932 (rank : 119) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
OLIG2_MOUSE
|
||||||
| NC score | 0.058208 (rank : 120) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9EQW6, Q9JKN4 | Gene names | Olig2 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2). | |||||
|
BHLH2_HUMAN
|
||||||
| NC score | 0.058043 (rank : 121) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
ID2_HUMAN
|
||||||
| NC score | 0.056832 (rank : 122) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02363 | Gene names | ID2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein inhibitor ID-2 (Inhibitor of DNA binding 2). | |||||
|
BHLH3_MOUSE
|
||||||
| NC score | 0.056521 (rank : 123) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
OLIG3_MOUSE
|
||||||
| NC score | 0.055870 (rank : 124) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PFG8, Q9EQW5 | Gene names | Olig3, Bhlhb7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oligodendrocyte transcription factor 3 (Oligo3) (Oligodendrocyte- specific bHLH transcription factor 3) (Basic helix-loop-helix domain- containing class B protein 7). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.055286 (rank : 125) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
OLIG3_HUMAN
|
||||||
| NC score | 0.055038 (rank : 126) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7RTU3, Q8N8Q0 | Gene names | OLIG3, BHLHB7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oligodendrocyte transcription factor 3 (Oligo3) (Basic helix-loop- helix domain-containing class B protein 7). | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.054905 (rank : 127) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
OLIG1_HUMAN
|
||||||
| NC score | 0.054883 (rank : 128) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TAK6, Q7RTS0 | Gene names | OLIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1). | |||||
|
OLIG1_MOUSE
|
||||||
| NC score | 0.054573 (rank : 129) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JKN5 | Gene names | Olig1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1) (Oligodendrocyte- specific bHLH transcription factor 1). | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.052266 (rank : 130) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
ID2_MOUSE
|
||||||
| NC score | 0.051398 (rank : 131) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P41136, O88604 | Gene names | Id2, Id-2, Idb2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein inhibitor ID-2 (Inhibitor of DNA binding 2). | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.051248 (rank : 132) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
TFE2_HUMAN
|
||||||
| NC score | 0.050357 (rank : 133) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P15923, P15883, Q14208, Q14635, Q14636, Q9UPI9 | Gene names | TCF3, E2A, ITF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Immunoglobulin transcription factor 1) (Transcription factor ITF-1) (Kappa-E2-binding factor). | |||||
|
CBX2_HUMAN
|
||||||
| NC score | 0.047900 (rank : 134) | θ value | 0.0252991 (rank : 32) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
GDIA_HUMAN
|
||||||
| NC score | 0.040669 (rank : 135) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31150, P50394, Q7Z2G6, Q7Z2G9, Q7Z2H5, Q7Z2I6 | Gene names | GDI1, OPHN2, RABGDIA, XAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rab GDP dissociation inhibitor alpha (Rab GDI alpha) (Guanosine diphosphate dissociation inhibitor 1) (GDI-1) (XAP-4) (Oligophrenin- 2). | |||||
|
GDIA_MOUSE
|
||||||
| NC score | 0.037586 (rank : 136) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P50396, Q8VHM3, Q91Y71, Q96CX5 | Gene names | Gdi1, Rabgdia | |||
|
Domain Architecture |

|
|||||
| Description | Rab GDP dissociation inhibitor alpha (Rab GDI alpha) (Guanosine diphosphate dissociation inhibitor 1) (GDI-1). | |||||
|
GDIB_MOUSE
|
||||||
| NC score | 0.037273 (rank : 137) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61598, Q541Z9, Q8C530, Q9D8M9 | Gene names | Gdi2, Gdi3 | |||
|
Domain Architecture |

|
|||||
| Description | Rab GDP dissociation inhibitor beta (Rab GDI beta) (Guanosine diphosphate dissociation inhibitor 2) (GDI-2) (GDI-3). | |||||
|
SDPR_MOUSE
|
||||||
| NC score | 0.033298 (rank : 138) | θ value | 0.47712 (rank : 57) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
SPIB_MOUSE
|
||||||
| NC score | 0.032528 (rank : 139) | θ value | 0.813845 (rank : 64) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.029409 (rank : 140) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.027703 (rank : 141) | θ value | 0.365318 (rank : 52) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
PODXL_HUMAN
|
||||||
| NC score | 0.027458 (rank : 142) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.026495 (rank : 143) | θ value | 0.163984 (rank : 45) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CSF1_HUMAN
|
||||||
| NC score | 0.023264 (rank : 144) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
GAK12_HUMAN
|
||||||
| NC score | 0.022012 (rank : 145) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
OSBP2_HUMAN
|
||||||
| NC score | 0.021909 (rank : 146) | θ value | 0.47712 (rank : 56) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q969R2, O60396, Q9BY96, Q9BZF0 | Gene names | OSBP2, KIAA1664, ORP4, OSBPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 2 (Oxysterol-binding protein-related protein 4) (OSBP-related protein 4) (ORP-4). | |||||
|
GAK6_HUMAN
|
||||||
| NC score | 0.021726 (rank : 147) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.021595 (rank : 148) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| NC score | 0.021588 (rank : 149) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.021519 (rank : 150) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
RL7_MOUSE
|
||||||
| NC score | 0.021425 (rank : 151) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P14148, Q80UR0 | Gene names | Rpl7 | |||
|
Domain Architecture |

|
|||||
| Description | 60S ribosomal protein L7. | |||||
|
GAK10_HUMAN
|
||||||
| NC score | 0.021065 (rank : 152) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.020880 (rank : 153) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
SMBP2_HUMAN
|
||||||
| NC score | 0.019593 (rank : 154) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.019533 (rank : 155) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
TEF_MOUSE
|
||||||
| NC score | 0.017371 (rank : 156) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLC6, Q3U426, Q3UGF4, Q3UM49, Q3URC8, Q6QHT6, Q8C6I0, Q8VD02 | Gene names | Tef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
TEF_HUMAN
|
||||||
| NC score | 0.017310 (rank : 157) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q10587, Q15729, Q7Z3J7, Q8IU94, Q96TG4 | Gene names | TEF | |||
|
Domain Architecture |

|
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.016695 (rank : 158) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.016564 (rank : 159) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
RHG05_HUMAN
|
||||||
| NC score | 0.015825 (rank : 160) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13017 | Gene names | ARHGAP5, RHOGAP5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
FGD5_HUMAN
|
||||||
| NC score | 0.015714 (rank : 161) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
EPN1_HUMAN
|
||||||
| NC score | 0.015490 (rank : 162) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
RHG05_MOUSE
|
||||||
| NC score | 0.015490 (rank : 163) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.015480 (rank : 164) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
DOCK6_HUMAN
|
||||||
| NC score | 0.015477 (rank : 165) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96HP0, Q9P2F2 | Gene names | DOCK6, KIAA1395 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 6. | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.015318 (rank : 166) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CEBPB_HUMAN
|
||||||
| NC score | 0.015256 (rank : 167) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17676, Q96IH2, Q9H4Z5 | Gene names | CEBPB, TCF5 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Nuclear factor NF- IL6) (Transcription factor 5). | |||||
|
CASP_HUMAN
|
||||||
| NC score | 0.015042 (rank : 168) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
SMC4_HUMAN
|
||||||
| NC score | 0.014371 (rank : 169) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.014294 (rank : 170) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.013005 (rank : 171) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
GATA6_HUMAN
|
||||||
| NC score | 0.012902 (rank : 172) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92908, P78327 | Gene names | GATA6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-6 (GATA-binding factor 6). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.012790 (rank : 173) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.012712 (rank : 174) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.012709 (rank : 175) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
SRCA_MOUSE
|
||||||
| NC score | 0.012508 (rank : 176) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.012505 (rank : 177) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
SNPC2_MOUSE
|
||||||
| NC score | 0.012224 (rank : 178) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91XA5, Q3UK17, Q8CBS9 | Gene names | Snapc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 2 (SNAPc subunit 2) (snRNA- activating protein complex 45 kDa subunit) (SNAPc 45 kDa subunit) (Small nuclear RNA-activating complex polypeptide 2). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.012193 (rank : 179) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
TTLL4_HUMAN
|
||||||
| NC score | 0.011904 (rank : 180) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14679, Q8WW29 | Gene names | TTLL4, KIAA0173 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
PEPL_MOUSE
|
||||||
| NC score | 0.010361 (rank : 181) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
ETV3_HUMAN
|
||||||
| NC score | 0.009200 (rank : 182) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.008525 (rank : 183) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
ALS2_HUMAN
|
||||||
| NC score | 0.006039 (rank : 184) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
KI21A_HUMAN
|
||||||
| NC score | 0.005884 (rank : 185) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.005660 (rank : 186) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
UN45B_MOUSE
|
||||||
| NC score | 0.005548 (rank : 187) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CGY6, Q5XG72, Q8BHC5, Q8BWK3 | Gene names | Unc45b, Cmya4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog B (UNC-45B). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.003311 (rank : 188) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
KCNC4_HUMAN
|
||||||
| NC score | 0.002199 (rank : 189) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
WNK4_HUMAN
|
||||||
| NC score | 0.000651 (rank : 190) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||