Please be patient as the page loads
|
MYOD1_HUMAN
|
||||||
| SwissProt Accessions | P15172 | Gene names | MYOD1, MYF3, MYOD | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1 (Myogenic factor 3) (Myf-3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MYOD1_HUMAN
|
||||||
| θ value | 7.02726e-162 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | P15172 | Gene names | MYOD1, MYF3, MYOD | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1 (Myogenic factor 3) (Myf-3). | |||||
|
MYOD1_MOUSE
|
||||||
| θ value | 6.62324e-136 (rank : 2) | NC score | 0.990038 (rank : 2) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P10085 | Gene names | Myod1, Myod | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1. | |||||
|
MYF5_HUMAN
|
||||||
| θ value | 2.75849e-49 (rank : 3) | NC score | 0.929813 (rank : 3) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P13349 | Gene names | MYF5 | |||
|
Domain Architecture |
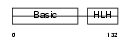
|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
MYF5_MOUSE
|
||||||
| θ value | 1.28137e-46 (rank : 4) | NC score | 0.928236 (rank : 4) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P24699, Q543W7 | Gene names | Myf5, Myf-5 | |||
|
Domain Architecture |
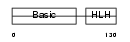
|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
MYOG_HUMAN
|
||||||
| θ value | 2.87452e-22 (rank : 5) | NC score | 0.830539 (rank : 7) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P15173 | Gene names | MYOG, MYF4 | |||
|
Domain Architecture |
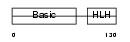
|
|||||
| Description | Myogenin (Myogenic factor 4) (Myf-4). | |||||
|
MYF6_HUMAN
|
||||||
| θ value | 5.42112e-21 (rank : 6) | NC score | 0.841764 (rank : 5) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P23409, Q53X80, Q6FHI9 | Gene names | MYF6 | |||
|
Domain Architecture |
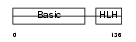
|
|||||
| Description | Myogenic factor 6 (Myf-6). | |||||
|
MYF6_MOUSE
|
||||||
| θ value | 5.42112e-21 (rank : 7) | NC score | 0.839812 (rank : 6) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P15375 | Gene names | Myf6, Myf-6 | |||
|
Domain Architecture |
|
|||||
| Description | Myogenic factor 6 (Myf-6) (Herculin). | |||||
|
MYOG_MOUSE
|
||||||
| θ value | 2.69047e-20 (rank : 8) | NC score | 0.817634 (rank : 8) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P12979 | Gene names | Myog | |||
|
Domain Architecture |
|
|||||
| Description | Myogenin (MYOD1-related protein). | |||||
|
LYL1_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 9) | NC score | 0.428755 (rank : 24) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P27792 | Gene names | Lyl1, Lyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
PTF1A_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 10) | NC score | 0.475751 (rank : 12) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7RTS3, Q9HC25 | Gene names | PTF1A, PTF1P48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
PTF1A_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 11) | NC score | 0.469964 (rank : 14) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9QX98, Q9QYF5, Q9QYF6 | Gene names | Ptf1a, Ptf1p48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
TWST1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 12) | NC score | 0.483314 (rank : 9) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q15672, Q92487, Q99804 | Gene names | TWIST1, TWIST | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (H-twist). | |||||
|
TWST1_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 13) | NC score | 0.483021 (rank : 10) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P26687 | Gene names | Twist1, Twist | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (M-twist). | |||||
|
TWST2_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 14) | NC score | 0.461231 (rank : 16) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8WVJ9 | Gene names | TWIST2, DERMO1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST2_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 15) | NC score | 0.461921 (rank : 15) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9D030 | Gene names | Twist2, Dermo1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TAL1_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 16) | NC score | 0.442877 (rank : 18) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P22091 | Gene names | Tal1, Scl, Tal-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein homolog (TAL-1 protein) (Stem cell protein). | |||||
|
TAL1_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 17) | NC score | 0.436519 (rank : 21) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P17542 | Gene names | TAL1, SCL, TCL5 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein (TAL-1 protein) (Stem cell protein) (T-cell leukemia/lymphoma-5 protein). | |||||
|
ASCL1_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 18) | NC score | 0.440392 (rank : 20) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q02067 | Gene names | Ascl1, Ash1, Mash-1, Mash1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (Mash-1). | |||||
|
LYL1_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 19) | NC score | 0.426637 (rank : 26) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P12980, O76102 | Gene names | LYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
NGN1_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 20) | NC score | 0.395509 (rank : 31) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P70660 | Gene names | Neurog1, Ath4c, Neurod3, Ngn, Ngn1 | |||
|
Domain Architecture |
|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein) (Helix-loop-helix protein mATH-4C) (MATH4C). | |||||
|
ASCL1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 21) | NC score | 0.435626 (rank : 22) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P50553, Q9BQ30 | Gene names | ASCL1, ASH1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (HASH1). | |||||
|
NGN1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 22) | NC score | 0.391611 (rank : 34) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q92886, Q96HE1 | Gene names | NEUROG1, NEUROD3, NGN, NGN1 | |||
|
Domain Architecture |
|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein). | |||||
|
FIGLA_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 23) | NC score | 0.478991 (rank : 11) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6QHK4 | Gene names | FIGLA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
NGN3_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 24) | NC score | 0.394350 (rank : 32) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70661 | Gene names | Neurog3, Ath4b, Atoh5, Ngn3 | |||
|
Domain Architecture |
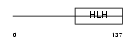
|
|||||
| Description | Neurogenin 3 (Atonal protein homolog 5) (Helix-loop-helix protein mATH-4B) (MATH4B). | |||||
|
TCF15_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 25) | NC score | 0.369950 (rank : 37) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q60756, Q60788 | Gene names | Tcf15, Bhlhec2 | |||
|
Domain Architecture |
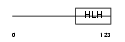
|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis) (Meso1). | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 26) | NC score | 0.032769 (rank : 104) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
FIGLA_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 27) | NC score | 0.472246 (rank : 13) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O55208 | Gene names | Figla | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
ASCL3_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 28) | NC score | 0.446245 (rank : 17) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JJR7 | Gene names | Ascl3, Mash3, Sgn1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 3 (bHLH transcriptional regulator Sgn-1) (Mash- 3). | |||||
|
SCX_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 29) | NC score | 0.401567 (rank : 27) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q64124 | Gene names | Scx | |||
|
Domain Architecture |
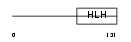
|
|||||
| Description | Basic helix-loop-helix transcription factor scleraxis. | |||||
|
TFE2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 30) | NC score | 0.227385 (rank : 63) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P15923, P15883, Q14208, Q14635, Q14636, Q9UPI9 | Gene names | TCF3, E2A, ITF1 | |||
|
Domain Architecture |
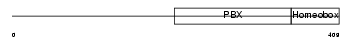
|
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Immunoglobulin transcription factor 1) (Transcription factor ITF-1) (Kappa-E2-binding factor). | |||||
|
HAND1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 31) | NC score | 0.373566 (rank : 36) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q64279, Q61099 | Gene names | Hand1, Ehand, Hxt, Thing1 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND) (Helix-loop-helix transcription factor expressed in extraembryonic mesoderm and trophoblast) (Thing-1) (Th1). | |||||
|
HAND2_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 32) | NC score | 0.397732 (rank : 28) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P61296, O95300, O95301, P97833 | Gene names | HAND2, DHAND | |||
|
Domain Architecture |
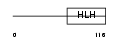
|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND). | |||||
|
HAND2_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 33) | NC score | 0.397732 (rank : 29) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q61039, Q61100 | Gene names | Hand2, Dhand, Hed, Thing2 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND) (Helix-loop-helix transcription factor expressed in embryo and deciduum) (Thing-2). | |||||
|
NDF2_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 34) | NC score | 0.340235 (rank : 41) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q15784, Q8TBI7, Q9UQC6 | Gene names | NEUROD2, NDRF | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
NDF2_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 35) | NC score | 0.340213 (rank : 42) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q62414, Q61952, Q925V5 | Gene names | Neurod2, Ndrf | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
ASCL3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 36) | NC score | 0.428639 (rank : 25) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NQ33, Q8WYQ6 | Gene names | ASCL3, HASH3, SGN1 | |||
|
Domain Architecture |
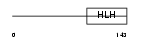
|
|||||
| Description | Achaete-scute homolog 3 (bHLH transcriptional regulator Sgn-1). | |||||
|
NGN3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 37) | NC score | 0.377704 (rank : 35) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y4Z2, Q9BY24 | Gene names | NEUROG3, NGN3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3. | |||||
|
ASCL2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 38) | NC score | 0.442649 (rank : 19) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O35885, Q9WUJ7 | Gene names | Ascl2, Mash2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash-2). | |||||
|
MNT_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 39) | NC score | 0.195534 (rank : 71) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 40) | NC score | 0.202725 (rank : 70) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
NDF1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 41) | NC score | 0.323415 (rank : 46) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
NDF1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 42) | NC score | 0.321584 (rank : 48) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
ASCL2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 43) | NC score | 0.432382 (rank : 23) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q99929, Q9UM68 | Gene names | ASCL2 | |||
|
Domain Architecture |
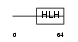
|
|||||
| Description | Achaete-scute homolog 2 (Mash2) (HASH2). | |||||
|
BHLH8_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 44) | NC score | 0.396835 (rank : 30) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q7RTS1 | Gene names | BHLHB8, MIST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
HTF4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 45) | NC score | 0.204368 (rank : 68) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99081 | Gene names | TCF12, HEB, HTF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4). | |||||
|
HTF4_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 46) | NC score | 0.211937 (rank : 64) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61286 | Gene names | Tcf12, Alf1, Meb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4) (Class A helix-loop-helix transcription factor ME1). | |||||
|
NDF4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 47) | NC score | 0.323349 (rank : 47) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9HD90 | Gene names | NEUROD4 | |||
|
Domain Architecture |
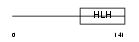
|
|||||
| Description | Neurogenic differentiation factor 4 (NeuroD4). | |||||
|
NDF4_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 48) | NC score | 0.324082 (rank : 45) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O09105 | Gene names | Neurod4, Ath3, Atoh3 | |||
|
Domain Architecture |
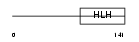
|
|||||
| Description | Neurogenic differentiation factor 4 (NeuroD4) (Atonal protein homolog 3) (Helix-loop-helix protein mATH-3) (MATH3). | |||||
|
TFE2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 49) | NC score | 0.211630 (rank : 65) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P15806, Q3U153, Q8CAH9, Q8VCY4, Q922S2, Q99MB8, Q9CYJ4 | Gene names | Tcf3, Alf2, Me2, Tcfe2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Transcription factor A1). | |||||
|
BHLH8_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 50) | NC score | 0.391885 (rank : 33) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
ITF2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 51) | NC score | 0.205454 (rank : 66) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P15884, Q15439, Q15440 | Gene names | TCF4, ITF2, SEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (SL3-3 enhancer factor 2) (SEF-2). | |||||
|
ITF2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 52) | NC score | 0.205233 (rank : 67) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60722, Q60721, Q62211, Q64072, Q80UE8 | Gene names | Tcf4, Itf2, Sef2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (MITF-2) (SL3-3 enhancer factor 2) (SEF-2) (Class A helix-loop-helix transcription factor ME2). | |||||
|
HAND1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 53) | NC score | 0.356914 (rank : 38) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O96004 | Gene names | HAND1, EHAND | |||
|
Domain Architecture |
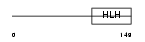
|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND). | |||||
|
ID1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 54) | NC score | 0.203476 (rank : 69) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P41134, O00651, O00652, Q16371, Q16377, Q5TE66, Q5TE67, Q969Z7, Q9H0Z5, Q9H109 | Gene names | ID1, ID | |||
|
Domain Architecture |
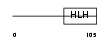
|
|||||
| Description | DNA-binding protein inhibitor ID-1 (Inhibitor of DNA binding 1). | |||||
|
NGN2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 55) | NC score | 0.348117 (rank : 39) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H2A3, Q8N416 | Gene names | NEUROG2, NGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogenin 2. | |||||
|
NGN2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 56) | NC score | 0.347666 (rank : 40) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P70447, P70237 | Gene names | Neurog2, Ath4a, Atoh4, Ngn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 2 (Atonal protein homolog 4) (Helix-loop-helix protein mATH-4A) (MATH4A). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.163984 (rank : 57) | NC score | 0.030874 (rank : 106) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
BAT3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 58) | NC score | 0.041186 (rank : 100) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
HES1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 59) | NC score | 0.114636 (rank : 86) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
HES1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 60) | NC score | 0.114725 (rank : 85) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
MYC_MOUSE
|
||||||
| θ value | 0.279714 (rank : 61) | NC score | 0.170834 (rank : 75) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
TFAP4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 62) | NC score | 0.306159 (rank : 50) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
ATF6B_MOUSE
|
||||||
| θ value | 0.365318 (rank : 63) | NC score | 0.036683 (rank : 101) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35451 | Gene names | Crebl1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 64) | NC score | 0.021234 (rank : 111) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | 0.813845 (rank : 65) | NC score | 0.036249 (rank : 102) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
MYOM1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 66) | NC score | 0.014318 (rank : 118) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62234 | Gene names | Myom1 | |||
|
Domain Architecture |

|
|||||
| Description | Myomesin-1 (Skelemin). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 67) | NC score | 0.018622 (rank : 114) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TTLL5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 68) | NC score | 0.027034 (rank : 108) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6EMB2, Q9P1V5, Q9UPZ4 | Gene names | TTLL5, KIAA0998, STAMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5 (SRC1 and TIF2-associated modulatory protein). | |||||
|
HES6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.074972 (rank : 95) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96HZ4, Q8N2J2, Q96T93, Q9P2S3 | Gene names | HES6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6) (C- HAIRY1). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.008676 (rank : 122) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
HES6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.068316 (rank : 97) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JHE6, Q3U2D7 | Gene names | Hes6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.026699 (rank : 109) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
TF3C2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.033308 (rank : 103) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WUA4, Q16632, Q9BWI7 | Gene names | GTF3C2, KIAA0011 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 2 (Transcription factor IIIC-subunit beta) (TF3C-beta) (TFIIIC 110 kDa subunit) (TFIIIC110). | |||||
|
CCNK_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.021275 (rank : 110) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.002754 (rank : 129) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
HES2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.069858 (rank : 96) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O54792 | Gene names | Hes2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
MYC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.133339 (rank : 84) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.015567 (rank : 116) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
R4RL2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.004546 (rank : 126) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7M6Z0, Q3UQ62 | Gene names | Rtn4rl2, Ngrl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4 receptor-like 2 precursor (Nogo-66 receptor homolog 1) (Nogo-66 receptor-related protein 2) (NgR2) (Nogo receptor-like 3). | |||||
|
RFX1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.012821 (rank : 119) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
RUSC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.014747 (rank : 117) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BVN2, Q9UPY4, Q9Y4T5 | Gene names | RUSC1, NESCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1 (New molecule containing SH3 at the carboxy-terminus) (Nesca). | |||||
|
TCF15_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.315400 (rank : 49) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q12870, Q9NQQ1 | Gene names | TCF15, BHLHEC2 | |||
|
Domain Architecture |
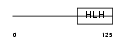
|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis). | |||||
|
TF3C2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.031008 (rank : 105) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BL74, Q3U7Z9, Q80XQ7, Q8BJJ4, Q8BLP3, Q8K1J1, Q9CSS0, Q9CVE6, Q9DBK6 | Gene names | Gtf3c2, Kiaa0011 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 2 (Transcription factor IIIC-subunit beta) (TF3C-beta) (TFIIIC 110 kDa subunit) (TFIIIC110). | |||||
|
HES4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.081039 (rank : 91) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
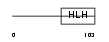
ID4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.162540 (rank : 79) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P47928, Q13005 | Gene names | ID4 | |||
|
Domain Architecture |
|
|||||
| Description | DNA-binding protein inhibitor ID-4 (Inhibitor of DNA binding 4). | |||||
|
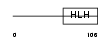
ID4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.158112 (rank : 82) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P41139 | Gene names | Id4, Id-4, Idb4 | |||
|
Domain Architecture |
|
|||||
| Description | DNA-binding protein inhibitor ID-4 (Inhibitor of DNA binding 4). | |||||
|
PRP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.018807 (rank : 113) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.008747 (rank : 121) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
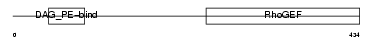
ARHG2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.006872 (rank : 124) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92974, O75142, Q15079 | Gene names | ARHGEF2, KIAA0651, LFP40 | |||
|
Domain Architecture |
|
|||||
| Description | Rho/Rac guanine nucleotide exchange factor 2 (GEF-H1 protein) (Proliferating cell nucleolar antigen p40). | |||||
|
CI005_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.019141 (rank : 112) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H330, Q5JTQ5, Q5SS43, Q6ZME3, Q8NDJ5, Q96CG6 | Gene names | C9orf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf5 (Protein CG-2). | |||||
|
MAX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.102553 (rank : 88) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MAX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.109453 (rank : 87) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
PDC6I_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.016845 (rank : 115) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
SOS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.008260 (rank : 123) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
TSP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.003832 (rank : 127) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35441 | Gene names | Thbs1, Tsp1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||
|
EVL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.009215 (rank : 120) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UI08, O95884, Q7Z522, Q8TBV1, Q9UF25, Q9UIC2 | Gene names | EVL, RNB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
NRG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.004561 (rank : 125) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14511 | Gene names | NRG2, NTAK | |||
|
Domain Architecture |

|
|||||
| Description | Pro-neuregulin-2, membrane-bound isoform precursor (Pro-NRG2) [Contains: Neuregulin-2 (NRG-2) (Neural- and thymus-derived activator for ERBB kinases) (NTAK) (Divergent of neuregulin-1) (DON-1)]. | |||||
|
R4RL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.003407 (rank : 128) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UN3, Q6X813 | Gene names | RTN4RL2, NGRH1, NGRL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4 receptor-like 2 precursor (Nogo-66 receptor homolog 1) (Nogo-66 receptor-related protein 2) (NgR2) (Nogo receptor-like 3). | |||||
|
TTLL5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.029773 (rank : 107) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | -0.000772 (rank : 130) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
ATOH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.251765 (rank : 56) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q92858 | Gene names | ATOH1, ATH1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein hATH-1). | |||||
|
ATOH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.258348 (rank : 55) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P48985 | Gene names | Atoh1, Ath1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein mATH-1) (MATH1). | |||||
|
BHLH4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.190497 (rank : 72) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NDY6 | Gene names | BHLHB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 4 (bHLHB4). | |||||
|
BHLH4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.184740 (rank : 73) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BGW3, Q71MB7 | Gene names | Bhlhb4, H20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 4 (bHLHB4). | |||||
|
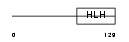
HEN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.298030 (rank : 51) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q02575 | Gene names | NHLH1, HEN1 | |||
|
Domain Architecture |
|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
HEN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.295472 (rank : 52) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q02576 | Gene names | Nhlh1, Hen1 | |||
|
Domain Architecture |
|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
HEN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.285973 (rank : 53) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q02577 | Gene names | NHLH2, HEN2 | |||
|
Domain Architecture |
|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
HEN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.284861 (rank : 54) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64221 | Gene names | Nhlh2, Hen2 | |||
|
Domain Architecture |
|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
ID1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.140133 (rank : 83) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P20067, Q61101, Q9D897 | Gene names | Id1, Id, Id-1, Idb1 | |||
|
Domain Architecture |
|
|||||
| Description | DNA-binding protein inhibitor ID-1 (Inhibitor of DNA binding 1). | |||||
|
ID2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.090633 (rank : 89) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02363 | Gene names | ID2 | |||
|
Domain Architecture |
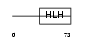
|
|||||
| Description | DNA-binding protein inhibitor ID-2 (Inhibitor of DNA binding 2). | |||||
|
ID2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.084435 (rank : 90) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P41136, O88604 | Gene names | Id2, Id-2, Idb2 | |||
|
Domain Architecture |
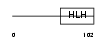
|
|||||
| Description | DNA-binding protein inhibitor ID-2 (Inhibitor of DNA binding 2). | |||||
|
MUSC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.240016 (rank : 58) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O60682, O75946, Q9BRE7 | Gene names | MSC, ABF1 | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Activated B-cell factor 1) (ABF-1). | |||||
|
MUSC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.244122 (rank : 57) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O88940 | Gene names | Msc, Myor | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Myogenic repressor). | |||||
|
MYCL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.059932 (rank : 98) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.054799 (rank : 99) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.076824 (rank : 93) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.076754 (rank : 94) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.077644 (rank : 92) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
NDF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.237306 (rank : 59) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96NK8, Q9H3H6 | Gene names | NEUROD6 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 6 (NeuroD6). | |||||
|
NDF6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.236262 (rank : 60) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P48986 | Gene names | Neurod6, Ath2, Atoh2, Nex1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 6 (NeuroD6) (Atonal protein homolog 2) (Helix-loop-helix protein mATH-2) (MATH2) (NEX-1 protein). | |||||
|
OLIG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.171656 (rank : 74) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TAK6, Q7RTS0 | Gene names | OLIG1 | |||
|
Domain Architecture |
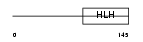
|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1). | |||||
|
OLIG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.168106 (rank : 78) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JKN5 | Gene names | Olig1 | |||
|
Domain Architecture |
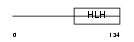
|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1) (Oligodendrocyte- specific bHLH transcription factor 1). | |||||
|
OLIG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.169667 (rank : 76) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13516, Q86X04, Q9NZ14 | Gene names | OLIG2, BHLHB1, PRKCBP2, RACK17 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2) (Basic helix-loop- helix protein class B 1) (Protein kinase C-binding protein RACK17) (Protein kinase C-binding protein 2). | |||||
|
OLIG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.168999 (rank : 77) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9EQW6, Q9JKN4 | Gene names | Olig2 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2). | |||||
|
OLIG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.160734 (rank : 81) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7RTU3, Q8N8Q0 | Gene names | OLIG3, BHLHB7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oligodendrocyte transcription factor 3 (Oligo3) (Basic helix-loop- helix domain-containing class B protein 7). | |||||
|
OLIG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.162395 (rank : 80) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6PFG8, Q9EQW5 | Gene names | Olig3, Bhlhb7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oligodendrocyte transcription factor 3 (Oligo3) (Oligodendrocyte- specific bHLH transcription factor 3) (Basic helix-loop-helix domain- containing class B protein 7). | |||||
|
TAL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.325025 (rank : 43) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q16559 | Gene names | TAL2 | |||
|
Domain Architecture |
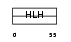
|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein (TAL-2 protein). | |||||
|
TAL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.324150 (rank : 44) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62282 | Gene names | Tal2 | |||
|
Domain Architecture |
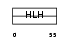
|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein homolog (TAL-2 protein). | |||||
|
TCF21_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.235285 (rank : 61) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O43680, O43545, Q9BZ14 | Gene names | TCF21, POD1 | |||
|
Domain Architecture |
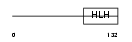
|
|||||
| Description | Transcription factor 21 (Podocyte-expressed 1) (Pod-1) (Epicardin) (Capsulin). | |||||
|
TCF21_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.234046 (rank : 62) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O35437, Q3U023 | Gene names | Tcf21, Pod1 | |||
|
Domain Architecture |
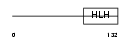
|
|||||
| Description | Transcription factor 21 (Podocyte-expressed 1) (Pod-1) (Epicardin) (Capsulin). | |||||
|
MYOD1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 7.02726e-162 (rank : 1) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | P15172 | Gene names | MYOD1, MYF3, MYOD | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1 (Myogenic factor 3) (Myf-3). | |||||
|
MYOD1_MOUSE
|
||||||
| NC score | 0.990038 (rank : 2) | θ value | 6.62324e-136 (rank : 2) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P10085 | Gene names | Myod1, Myod | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1. | |||||
|
MYF5_HUMAN
|
||||||
| NC score | 0.929813 (rank : 3) | θ value | 2.75849e-49 (rank : 3) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P13349 | Gene names | MYF5 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
MYF5_MOUSE
|
||||||
| NC score | 0.928236 (rank : 4) | θ value | 1.28137e-46 (rank : 4) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P24699, Q543W7 | Gene names | Myf5, Myf-5 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
MYF6_HUMAN
|
||||||
| NC score | 0.841764 (rank : 5) | θ value | 5.42112e-21 (rank : 6) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P23409, Q53X80, Q6FHI9 | Gene names | MYF6 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 6 (Myf-6). | |||||
|
MYF6_MOUSE
|
||||||
| NC score | 0.839812 (rank : 6) | θ value | 5.42112e-21 (rank : 7) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P15375 | Gene names | Myf6, Myf-6 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 6 (Myf-6) (Herculin). | |||||
|
MYOG_HUMAN
|
||||||
| NC score | 0.830539 (rank : 7) | θ value | 2.87452e-22 (rank : 5) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P15173 | Gene names | MYOG, MYF4 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenin (Myogenic factor 4) (Myf-4). | |||||
|
MYOG_MOUSE
|
||||||
| NC score | 0.817634 (rank : 8) | θ value | 2.69047e-20 (rank : 8) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P12979 | Gene names | Myog | |||
|
Domain Architecture |

|
|||||
| Description | Myogenin (MYOD1-related protein). | |||||
|
TWST1_HUMAN
|
||||||
| NC score | 0.483314 (rank : 9) | θ value | 0.000121331 (rank : 12) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q15672, Q92487, Q99804 | Gene names | TWIST1, TWIST | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (H-twist). | |||||
|
TWST1_MOUSE
|
||||||
| NC score | 0.483021 (rank : 10) | θ value | 0.000121331 (rank : 13) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P26687 | Gene names | Twist1, Twist | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (M-twist). | |||||
|
FIGLA_HUMAN
|
||||||
| NC score | 0.478991 (rank : 11) | θ value | 0.00175202 (rank : 23) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6QHK4 | Gene names | FIGLA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
PTF1A_HUMAN
|
||||||
| NC score | 0.475751 (rank : 12) | θ value | 0.000121331 (rank : 10) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7RTS3, Q9HC25 | Gene names | PTF1A, PTF1P48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
FIGLA_MOUSE
|
||||||
| NC score | 0.472246 (rank : 13) | θ value | 0.00298849 (rank : 27) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O55208 | Gene names | Figla | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
PTF1A_MOUSE
|
||||||
| NC score | 0.469964 (rank : 14) | θ value | 0.000121331 (rank : 11) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9QX98, Q9QYF5, Q9QYF6 | Gene names | Ptf1a, Ptf1p48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
TWST2_MOUSE
|
||||||
| NC score | 0.461921 (rank : 15) | θ value | 0.000270298 (rank : 15) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9D030 | Gene names | Twist2, Dermo1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST2_HUMAN
|
||||||
| NC score | 0.461231 (rank : 16) | θ value | 0.000270298 (rank : 14) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8WVJ9 | Gene names | TWIST2, DERMO1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
ASCL3_MOUSE
|
||||||
| NC score | 0.446245 (rank : 17) | θ value | 0.00665767 (rank : 28) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9JJR7 | Gene names | Ascl3, Mash3, Sgn1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 3 (bHLH transcriptional regulator Sgn-1) (Mash- 3). | |||||
|
TAL1_MOUSE
|
||||||
| NC score | 0.442877 (rank : 18) | θ value | 0.00035302 (rank : 16) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P22091 | Gene names | Tal1, Scl, Tal-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein homolog (TAL-1 protein) (Stem cell protein). | |||||
|
ASCL2_MOUSE
|
||||||
| NC score | 0.442649 (rank : 19) | θ value | 0.0330416 (rank : 38) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O35885, Q9WUJ7 | Gene names | Ascl2, Mash2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash-2). | |||||
|
ASCL1_MOUSE
|
||||||
| NC score | 0.440392 (rank : 20) | θ value | 0.00102713 (rank : 18) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q02067 | Gene names | Ascl1, Ash1, Mash-1, Mash1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (Mash-1). | |||||
|
TAL1_HUMAN
|
||||||
| NC score | 0.436519 (rank : 21) | θ value | 0.000602161 (rank : 17) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P17542 | Gene names | TAL1, SCL, TCL5 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein (TAL-1 protein) (Stem cell protein) (T-cell leukemia/lymphoma-5 protein). | |||||
|
ASCL1_HUMAN
|
||||||
| NC score | 0.435626 (rank : 22) | θ value | 0.00134147 (rank : 21) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P50553, Q9BQ30 | Gene names | ASCL1, ASH1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (HASH1). | |||||
|
ASCL2_HUMAN
|
||||||
| NC score | 0.432382 (rank : 23) | θ value | 0.0431538 (rank : 43) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q99929, Q9UM68 | Gene names | ASCL2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash2) (HASH2). | |||||
|
LYL1_MOUSE
|
||||||
| NC score | 0.428755 (rank : 24) | θ value | 7.1131e-05 (rank : 9) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P27792 | Gene names | Lyl1, Lyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
ASCL3_HUMAN
|
||||||
| NC score | 0.428639 (rank : 25) | θ value | 0.0252991 (rank : 36) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NQ33, Q8WYQ6 | Gene names | ASCL3, HASH3, SGN1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 3 (bHLH transcriptional regulator Sgn-1). | |||||
|
LYL1_HUMAN
|
||||||
| NC score | 0.426637 (rank : 26) | θ value | 0.00102713 (rank : 19) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P12980, O76102 | Gene names | LYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
SCX_MOUSE
|
||||||
| NC score | 0.401567 (rank : 27) | θ value | 0.00869519 (rank : 29) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q64124 | Gene names | Scx | |||
|
Domain Architecture |

|
|||||
| Description | Basic helix-loop-helix transcription factor scleraxis. | |||||
|
HAND2_HUMAN
|
||||||
| NC score | 0.397732 (rank : 28) | θ value | 0.0148317 (rank : 32) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P61296, O95300, O95301, P97833 | Gene names | HAND2, DHAND | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND). | |||||
|
HAND2_MOUSE
|
||||||
| NC score | 0.397732 (rank : 29) | θ value | 0.0148317 (rank : 33) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q61039, Q61100 | Gene names | Hand2, Dhand, Hed, Thing2 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND) (Helix-loop-helix transcription factor expressed in embryo and deciduum) (Thing-2). | |||||
|
BHLH8_HUMAN
|
||||||
| NC score | 0.396835 (rank : 30) | θ value | 0.0431538 (rank : 44) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q7RTS1 | Gene names | BHLHB8, MIST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
NGN1_MOUSE
|
||||||
| NC score | 0.395509 (rank : 31) | θ value | 0.00102713 (rank : 20) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P70660 | Gene names | Neurog1, Ath4c, Neurod3, Ngn, Ngn1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein) (Helix-loop-helix protein mATH-4C) (MATH4C). | |||||
|
NGN3_MOUSE
|
||||||
| NC score | 0.394350 (rank : 32) | θ value | 0.00175202 (rank : 24) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70661 | Gene names | Neurog3, Ath4b, Atoh5, Ngn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3 (Atonal protein homolog 5) (Helix-loop-helix protein mATH-4B) (MATH4B). | |||||
|
BHLH8_MOUSE
|
||||||
| NC score | 0.391885 (rank : 33) | θ value | 0.0961366 (rank : 50) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
NGN1_HUMAN
|
||||||
| NC score | 0.391611 (rank : 34) | θ value | 0.00134147 (rank : 22) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q92886, Q96HE1 | Gene names | NEUROG1, NEUROD3, NGN, NGN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein). | |||||
|
NGN3_HUMAN
|
||||||
| NC score | 0.377704 (rank : 35) | θ value | 0.0252991 (rank : 37) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y4Z2, Q9BY24 | Gene names | NEUROG3, NGN3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3. | |||||
|
HAND1_MOUSE
|
||||||
| NC score | 0.373566 (rank : 36) | θ value | 0.0113563 (rank : 31) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q64279, Q61099 | Gene names | Hand1, Ehand, Hxt, Thing1 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND) (Helix-loop-helix transcription factor expressed in extraembryonic mesoderm and trophoblast) (Thing-1) (Th1). | |||||
|
TCF15_MOUSE
|
||||||
| NC score | 0.369950 (rank : 37) | θ value | 0.00175202 (rank : 25) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q60756, Q60788 | Gene names | Tcf15, Bhlhec2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis) (Meso1). | |||||
|
HAND1_HUMAN
|
||||||
| NC score | 0.356914 (rank : 38) | θ value | 0.125558 (rank : 53) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O96004 | Gene names | HAND1, EHAND | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND). | |||||
|
NGN2_HUMAN
|
||||||
| NC score | 0.348117 (rank : 39) | θ value | 0.125558 (rank : 55) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H2A3, Q8N416 | Gene names | NEUROG2, NGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogenin 2. | |||||
|
NGN2_MOUSE
|
||||||
| NC score | 0.347666 (rank : 40) | θ value | 0.125558 (rank : 56) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P70447, P70237 | Gene names | Neurog2, Ath4a, Atoh4, Ngn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 2 (Atonal protein homolog 4) (Helix-loop-helix protein mATH-4A) (MATH4A). | |||||
|
NDF2_HUMAN
|
||||||
| NC score | 0.340235 (rank : 41) | θ value | 0.0148317 (rank : 34) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q15784, Q8TBI7, Q9UQC6 | Gene names | NEUROD2, NDRF | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
NDF2_MOUSE
|
||||||
| NC score | 0.340213 (rank : 42) | θ value | 0.0148317 (rank : 35) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q62414, Q61952, Q925V5 | Gene names | Neurod2, Ndrf | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
TAL2_HUMAN
|
||||||
| NC score | 0.325025 (rank : 43) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q16559 | Gene names | TAL2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein (TAL-2 protein). | |||||
|
TAL2_MOUSE
|
||||||
| NC score | 0.324150 (rank : 44) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62282 | Gene names | Tal2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein homolog (TAL-2 protein). | |||||
|
NDF4_MOUSE
|
||||||
| NC score | 0.324082 (rank : 45) | θ value | 0.0736092 (rank : 48) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O09105 | Gene names | Neurod4, Ath3, Atoh3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 4 (NeuroD4) (Atonal protein homolog 3) (Helix-loop-helix protein mATH-3) (MATH3). | |||||
|
NDF1_HUMAN
|
||||||
| NC score | 0.323415 (rank : 46) | θ value | 0.0330416 (rank : 41) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
NDF4_HUMAN
|
||||||
| NC score | 0.323349 (rank : 47) | θ value | 0.0736092 (rank : 47) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9HD90 | Gene names | NEUROD4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 4 (NeuroD4). | |||||
|
NDF1_MOUSE
|
||||||
| NC score | 0.321584 (rank : 48) | θ value | 0.0330416 (rank : 42) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
TCF15_HUMAN
|
||||||
| NC score | 0.315400 (rank : 49) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q12870, Q9NQQ1 | Gene names | TCF15, BHLHEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis). | |||||
|
TFAP4_HUMAN
|
||||||
| NC score | 0.306159 (rank : 50) | θ value | 0.279714 (rank : 62) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
HEN1_HUMAN
|
||||||
| NC score | 0.298030 (rank : 51) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q02575 | Gene names | NHLH1, HEN1 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
HEN1_MOUSE
|
||||||
| NC score | 0.295472 (rank : 52) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q02576 | Gene names | Nhlh1, Hen1 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
HEN2_HUMAN
|
||||||
| NC score | 0.285973 (rank : 53) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q02577 | Gene names | NHLH2, HEN2 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
HEN2_MOUSE
|
||||||
| NC score | 0.284861 (rank : 54) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q64221 | Gene names | Nhlh2, Hen2 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
ATOH1_MOUSE
|
||||||
| NC score | 0.258348 (rank : 55) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P48985 | Gene names | Atoh1, Ath1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein mATH-1) (MATH1). | |||||
|
ATOH1_HUMAN
|
||||||
| NC score | 0.251765 (rank : 56) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q92858 | Gene names | ATOH1, ATH1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein hATH-1). | |||||
|
MUSC_MOUSE
|
||||||
| NC score | 0.244122 (rank : 57) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O88940 | Gene names | Msc, Myor | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Myogenic repressor). | |||||
|
MUSC_HUMAN
|
||||||
| NC score | 0.240016 (rank : 58) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O60682, O75946, Q9BRE7 | Gene names | MSC, ABF1 | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Activated B-cell factor 1) (ABF-1). | |||||
|
NDF6_HUMAN
|
||||||
| NC score | 0.237306 (rank : 59) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96NK8, Q9H3H6 | Gene names | NEUROD6 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 6 (NeuroD6). | |||||
|
NDF6_MOUSE
|
||||||
| NC score | 0.236262 (rank : 60) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P48986 | Gene names | Neurod6, Ath2, Atoh2, Nex1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 6 (NeuroD6) (Atonal protein homolog 2) (Helix-loop-helix protein mATH-2) (MATH2) (NEX-1 protein). | |||||
|
TCF21_HUMAN
|
||||||
| NC score | 0.235285 (rank : 61) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O43680, O43545, Q9BZ14 | Gene names | TCF21, POD1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 21 (Podocyte-expressed 1) (Pod-1) (Epicardin) (Capsulin). | |||||
|
TCF21_MOUSE
|
||||||
| NC score | 0.234046 (rank : 62) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O35437, Q3U023 | Gene names | Tcf21, Pod1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 21 (Podocyte-expressed 1) (Pod-1) (Epicardin) (Capsulin). | |||||
|
TFE2_HUMAN
|
||||||
| NC score | 0.227385 (rank : 63) | θ value | 0.00869519 (rank : 30) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P15923, P15883, Q14208, Q14635, Q14636, Q9UPI9 | Gene names | TCF3, E2A, ITF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Immunoglobulin transcription factor 1) (Transcription factor ITF-1) (Kappa-E2-binding factor). | |||||
|
HTF4_MOUSE
|
||||||
| NC score | 0.211937 (rank : 64) | θ value | 0.0736092 (rank : 46) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61286 | Gene names | Tcf12, Alf1, Meb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4) (Class A helix-loop-helix transcription factor ME1). | |||||
|
TFE2_MOUSE
|
||||||
| NC score | 0.211630 (rank : 65) | θ value | 0.0736092 (rank : 49) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P15806, Q3U153, Q8CAH9, Q8VCY4, Q922S2, Q99MB8, Q9CYJ4 | Gene names | Tcf3, Alf2, Me2, Tcfe2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Transcription factor A1). | |||||
|
ITF2_HUMAN
|
||||||
| NC score | 0.205454 (rank : 66) | θ value | 0.0961366 (rank : 51) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P15884, Q15439, Q15440 | Gene names | TCF4, ITF2, SEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (SL3-3 enhancer factor 2) (SEF-2). | |||||
|
ITF2_MOUSE
|
||||||
| NC score | 0.205233 (rank : 67) | θ value | 0.0961366 (rank : 52) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60722, Q60721, Q62211, Q64072, Q80UE8 | Gene names | Tcf4, Itf2, Sef2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (MITF-2) (SL3-3 enhancer factor 2) (SEF-2) (Class A helix-loop-helix transcription factor ME2). | |||||
|
HTF4_HUMAN
|
||||||
| NC score | 0.204368 (rank : 68) | θ value | 0.0736092 (rank : 45) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99081 | Gene names | TCF12, HEB, HTF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4). | |||||
|
ID1_HUMAN
|
||||||
| NC score | 0.203476 (rank : 69) | θ value | 0.125558 (rank : 54) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P41134, O00651, O00652, Q16371, Q16377, Q5TE66, Q5TE67, Q969Z7, Q9H0Z5, Q9H109 | Gene names | ID1, ID | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein inhibitor ID-1 (Inhibitor of DNA binding 1). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.202725 (rank : 70) | θ value | 0.0330416 (rank : 40) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.195534 (rank : 71) | θ value | 0.0330416 (rank : 39) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
BHLH4_HUMAN
|
||||||
| NC score | 0.190497 (rank : 72) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NDY6 | Gene names | BHLHB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 4 (bHLHB4). | |||||
|
BHLH4_MOUSE
|
||||||
| NC score | 0.184740 (rank : 73) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BGW3, Q71MB7 | Gene names | Bhlhb4, H20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 4 (bHLHB4). | |||||
|
OLIG1_HUMAN
|
||||||
| NC score | 0.171656 (rank : 74) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TAK6, Q7RTS0 | Gene names | OLIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1). | |||||
|
MYC_MOUSE
|
||||||
| NC score | 0.170834 (rank : 75) | θ value | 0.279714 (rank : 61) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
OLIG2_HUMAN
|
||||||
| NC score | 0.169667 (rank : 76) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13516, Q86X04, Q9NZ14 | Gene names | OLIG2, BHLHB1, PRKCBP2, RACK17 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2) (Basic helix-loop- helix protein class B 1) (Protein kinase C-binding protein RACK17) (Protein kinase C-binding protein 2). | |||||
|
OLIG2_MOUSE
|
||||||
| NC score | 0.168999 (rank : 77) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9EQW6, Q9JKN4 | Gene names | Olig2 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2). | |||||
|
OLIG1_MOUSE
|
||||||
| NC score | 0.168106 (rank : 78) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JKN5 | Gene names | Olig1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1) (Oligodendrocyte- specific bHLH transcription factor 1). | |||||
|
ID4_HUMAN
|
||||||
| NC score | 0.162540 (rank : 79) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P47928, Q13005 | Gene names | ID4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein inhibitor ID-4 (Inhibitor of DNA binding 4). | |||||
|
OLIG3_MOUSE
|
||||||
| NC score | 0.162395 (rank : 80) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6PFG8, Q9EQW5 | Gene names | Olig3, Bhlhb7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oligodendrocyte transcription factor 3 (Oligo3) (Oligodendrocyte- specific bHLH transcription factor 3) (Basic helix-loop-helix domain- containing class B protein 7). | |||||
|
OLIG3_HUMAN
|
||||||
| NC score | 0.160734 (rank : 81) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7RTU3, Q8N8Q0 | Gene names | OLIG3, BHLHB7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oligodendrocyte transcription factor 3 (Oligo3) (Basic helix-loop- helix domain-containing class B protein 7). | |||||
|
ID4_MOUSE
|
||||||
| NC score | 0.158112 (rank : 82) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P41139 | Gene names | Id4, Id-4, Idb4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein inhibitor ID-4 (Inhibitor of DNA binding 4). | |||||
|
ID1_MOUSE
|
||||||
| NC score | 0.140133 (rank : 83) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P20067, Q61101, Q9D897 | Gene names | Id1, Id, Id-1, Idb1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein inhibitor ID-1 (Inhibitor of DNA binding 1). | |||||
|
MYC_HUMAN
|
||||||
| NC score | 0.133339 (rank : 84) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
HES1_MOUSE
|
||||||
| NC score | 0.114725 (rank : 85) | θ value | 0.21417 (rank : 60) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
HES1_HUMAN
|
||||||
| NC score | 0.114636 (rank : 86) | θ value | 0.21417 (rank : 59) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
MAX_MOUSE
|
||||||
| NC score | 0.109453 (rank : 87) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MAX_HUMAN
|
||||||
| NC score | 0.102553 (rank : 88) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
ID2_HUMAN
|
||||||
| NC score | 0.090633 (rank : 89) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02363 | Gene names | ID2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein inhibitor ID-2 (Inhibitor of DNA binding 2). | |||||
|
ID2_MOUSE
|
||||||
| NC score | 0.084435 (rank : 90) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P41136, O88604 | Gene names | Id2, Id-2, Idb2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein inhibitor ID-2 (Inhibitor of DNA binding 2). | |||||
|
HES4_HUMAN
|
||||||
| NC score | 0.081039 (rank : 91) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
MYCS_MOUSE
|
||||||
| NC score | 0.077644 (rank : 92) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
MYCN_HUMAN
|
||||||
| NC score | 0.076824 (rank : 93) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCN_MOUSE
|
||||||
| NC score | 0.076754 (rank : 94) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
HES6_HUMAN
|
||||||
| NC score | 0.074972 (rank : 95) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96HZ4, Q8N2J2, Q96T93, Q9P2S3 | Gene names | HES6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6) (C- HAIRY1). | |||||
|
HES2_MOUSE
|
||||||
| NC score | 0.069858 (rank : 96) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O54792 | Gene names | Hes2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HES6_MOUSE
|
||||||
| NC score | 0.068316 (rank : 97) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JHE6, Q3U2D7 | Gene names | Hes6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6). | |||||
|
MYCL1_HUMAN
|
||||||
| NC score | 0.059932 (rank : 98) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL1_MOUSE
|
||||||
| NC score | 0.054799 (rank : 99) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
BAT3_MOUSE
|
||||||
| NC score | 0.041186 (rank : 100) | θ value | 0.21417 (rank : 58) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
ATF6B_MOUSE
|
||||||
| NC score | 0.036683 (rank : 101) | θ value | 0.365318 (rank : 63) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35451 | Gene names | Crebl1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.036249 (rank : 102) | θ value | 0.813845 (rank : 65) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
TF3C2_HUMAN
|
||||||
| NC score | 0.033308 (rank : 103) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WUA4, Q16632, Q9BWI7 | Gene names | GTF3C2, KIAA0011 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 2 (Transcription factor IIIC-subunit beta) (TF3C-beta) (TFIIIC 110 kDa subunit) (TFIIIC110). | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.032769 (rank : 104) | θ value | 0.00228821 (rank : 26) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
TF3C2_MOUSE
|
||||||
| NC score | 0.031008 (rank : 105) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BL74, Q3U7Z9, Q80XQ7, Q8BJJ4, Q8BLP3, Q8K1J1, Q9CSS0, Q9CVE6, Q9DBK6 | Gene names | Gtf3c2, Kiaa0011 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 2 (Transcription factor IIIC-subunit beta) (TF3C-beta) (TFIIIC 110 kDa subunit) (TFIIIC110). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.030874 (rank : 106) | θ value | 0.163984 (rank : 57) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
TTLL5_MOUSE
|
||||||
| NC score | 0.029773 (rank : 107) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
TTLL5_HUMAN
|
||||||
| NC score | 0.027034 (rank : 108) | θ value | 0.813845 (rank : 68) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6EMB2, Q9P1V5, Q9UPZ4 | Gene names | TTLL5, KIAA0998, STAMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5 (SRC1 and TIF2-associated modulatory protein). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.026699 (rank : 109) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.021275 (rank : 110) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.021234 (rank : 111) | θ value | 0.365318 (rank : 64) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
CI005_HUMAN
|
||||||
| NC score | 0.019141 (rank : 112) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H330, Q5JTQ5, Q5SS43, Q6ZME3, Q8NDJ5, Q96CG6 | Gene names | C9orf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf5 (Protein CG-2). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.018807 (rank : 113) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.018622 (rank : 114) | θ value | 0.813845 (rank : 67) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PDC6I_MOUSE
|
||||||
| NC score | 0.016845 (rank : 115) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.015567 (rank : 116) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
RUSC1_HUMAN
|
||||||
| NC score | 0.014747 (rank : 117) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BVN2, Q9UPY4, Q9Y4T5 | Gene names | RUSC1, NESCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1 (New molecule containing SH3 at the carboxy-terminus) (Nesca). | |||||
|
MYOM1_MOUSE
|
||||||
| NC score | 0.014318 (rank : 118) | θ value | 0.813845 (rank : 66) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62234 | Gene names | Myom1 | |||
|
Domain Architecture |

|
|||||
| Description | Myomesin-1 (Skelemin). | |||||
|
RFX1_MOUSE
|
||||||
| NC score | 0.012821 (rank : 119) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
EVL_HUMAN
|
||||||
| NC score | 0.009215 (rank : 120) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UI08, O95884, Q7Z522, Q8TBV1, Q9UF25, Q9UIC2 | Gene names | EVL, RNB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.008747 (rank : 121) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.008676 (rank : 122) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
SOS1_MOUSE
|
||||||
| NC score | 0.008260 (rank : 123) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
ARHG2_HUMAN
|
||||||
| NC score | 0.006872 (rank : 124) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92974, O75142, Q15079 | Gene names | ARHGEF2, KIAA0651, LFP40 | |||
|
Domain Architecture |

|
|||||
| Description | Rho/Rac guanine nucleotide exchange factor 2 (GEF-H1 protein) (Proliferating cell nucleolar antigen p40). | |||||
|
NRG2_HUMAN
|
||||||
| NC score | 0.004561 (rank : 125) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14511 | Gene names | NRG2, NTAK | |||
|
Domain Architecture |

|
|||||
| Description | Pro-neuregulin-2, membrane-bound isoform precursor (Pro-NRG2) [Contains: Neuregulin-2 (NRG-2) (Neural- and thymus-derived activator for ERBB kinases) (NTAK) (Divergent of neuregulin-1) (DON-1)]. | |||||
|
R4RL2_MOUSE
|
||||||
| NC score | 0.004546 (rank : 126) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7M6Z0, Q3UQ62 | Gene names | Rtn4rl2, Ngrl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4 receptor-like 2 precursor (Nogo-66 receptor homolog 1) (Nogo-66 receptor-related protein 2) (NgR2) (Nogo receptor-like 3). | |||||
|
TSP1_MOUSE
|
||||||
| NC score | 0.003832 (rank : 127) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35441 | Gene names | Thbs1, Tsp1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||
|
R4RL2_HUMAN
|
||||||
| NC score | 0.003407 (rank : 128) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UN3, Q6X813 | Gene names | RTN4RL2, NGRH1, NGRL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4 receptor-like 2 precursor (Nogo-66 receptor homolog 1) (Nogo-66 receptor-related protein 2) (NgR2) (Nogo receptor-like 3). | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.002754 (rank : 129) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | -0.000772 (rank : 130) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||