Please be patient as the page loads
|
ITF2_HUMAN
|
||||||
| SwissProt Accessions | P15884, Q15439, Q15440 | Gene names | TCF4, ITF2, SEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (SL3-3 enhancer factor 2) (SEF-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HTF4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.963098 (rank : 4) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q99081 | Gene names | TCF12, HEB, HTF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4). | |||||
|
HTF4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.964384 (rank : 3) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q61286 | Gene names | Tcf12, Alf1, Meb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4) (Class A helix-loop-helix transcription factor ME1). | |||||
|
ITF2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 168 | |
| SwissProt Accessions | P15884, Q15439, Q15440 | Gene names | TCF4, ITF2, SEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (SL3-3 enhancer factor 2) (SEF-2). | |||||
|
ITF2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.991501 (rank : 2) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 130 | |
| SwissProt Accessions | Q60722, Q60721, Q62211, Q64072, Q80UE8 | Gene names | Tcf4, Itf2, Sef2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (MITF-2) (SL3-3 enhancer factor 2) (SEF-2) (Class A helix-loop-helix transcription factor ME2). | |||||
|
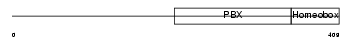
TFE2_HUMAN
|
||||||
| θ value | 3.06949e-141 (rank : 5) | NC score | 0.946189 (rank : 6) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P15923, P15883, Q14208, Q14635, Q14636, Q9UPI9 | Gene names | TCF3, E2A, ITF1 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Immunoglobulin transcription factor 1) (Transcription factor ITF-1) (Kappa-E2-binding factor). | |||||
|
TFE2_MOUSE
|
||||||
| θ value | 4.15819e-130 (rank : 6) | NC score | 0.946347 (rank : 5) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P15806, Q3U153, Q8CAH9, Q8VCY4, Q922S2, Q99MB8, Q9CYJ4 | Gene names | Tcf3, Alf2, Me2, Tcfe2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Transcription factor A1). | |||||
|
HORN_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 7) | NC score | 0.117388 (rank : 49) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 8) | NC score | 0.093258 (rank : 67) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
HORN_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 9) | NC score | 0.125766 (rank : 47) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
ATOH1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 10) | NC score | 0.240755 (rank : 7) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P48985 | Gene names | Atoh1, Ath1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein mATH-1) (MATH1). | |||||
|
BHLH8_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 11) | NC score | 0.212558 (rank : 12) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
CDKL5_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 12) | NC score | 0.015357 (rank : 157) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
PPRB_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 13) | NC score | 0.080152 (rank : 77) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
ATOH1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 14) | NC score | 0.216088 (rank : 11) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92858 | Gene names | ATOH1, ATH1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein hATH-1). | |||||
|
EGR1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 15) | NC score | 0.012675 (rank : 167) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
NDF2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 16) | NC score | 0.194944 (rank : 19) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q15784, Q8TBI7, Q9UQC6 | Gene names | NEUROD2, NDRF | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
NDF2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 17) | NC score | 0.194568 (rank : 20) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62414, Q61952, Q925V5 | Gene names | Neurod2, Ndrf | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
PPRB_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 18) | NC score | 0.075185 (rank : 82) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
LRRF2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 19) | NC score | 0.037505 (rank : 108) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
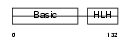
MYF5_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 20) | NC score | 0.223829 (rank : 10) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P13349 | Gene names | MYF5 | |||
|
Domain Architecture |
|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
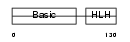
MYF5_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 21) | NC score | 0.226738 (rank : 8) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P24699, Q543W7 | Gene names | Myf5, Myf-5 | |||
|
Domain Architecture |
|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
BHLH8_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 22) | NC score | 0.190246 (rank : 22) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q7RTS1 | Gene names | BHLHB8, MIST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
FIGLA_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 23) | NC score | 0.224488 (rank : 9) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O55208 | Gene names | Figla | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
TWST1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 24) | NC score | 0.199023 (rank : 15) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15672, Q92487, Q99804 | Gene names | TWIST1, TWIST | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (H-twist). | |||||
|
TWST1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 25) | NC score | 0.198711 (rank : 16) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P26687 | Gene names | Twist1, Twist | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (M-twist). | |||||
|
MYOD1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 26) | NC score | 0.205454 (rank : 14) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P15172 | Gene names | MYOD1, MYF3, MYOD | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1 (Myogenic factor 3) (Myf-3). | |||||
|
MYOD1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 27) | NC score | 0.207464 (rank : 13) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P10085 | Gene names | Myod1, Myod | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1. | |||||
|
NDF6_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 28) | NC score | 0.169788 (rank : 34) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96NK8, Q9H3H6 | Gene names | NEUROD6 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 6 (NeuroD6). | |||||
|
NDF6_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 29) | NC score | 0.169849 (rank : 33) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P48986 | Gene names | Neurod6, Ath2, Atoh2, Nex1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 6 (NeuroD6) (Atonal protein homolog 2) (Helix-loop-helix protein mATH-2) (MATH2) (NEX-1 protein). | |||||
|
TWST2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 30) | NC score | 0.196458 (rank : 18) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8WVJ9 | Gene names | TWIST2, DERMO1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 31) | NC score | 0.196661 (rank : 17) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9D030 | Gene names | Twist2, Dermo1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
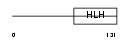
HEN2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 32) | NC score | 0.185513 (rank : 25) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02577 | Gene names | NHLH2, HEN2 | |||
|
Domain Architecture |
|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
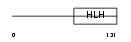
HEN2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 33) | NC score | 0.185398 (rank : 26) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q64221 | Gene names | Nhlh2, Hen2 | |||
|
Domain Architecture |
|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
NDF1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 34) | NC score | 0.178979 (rank : 30) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
CI039_HUMAN
|
||||||
| θ value | 0.163984 (rank : 35) | NC score | 0.027476 (rank : 122) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
NDF1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 36) | NC score | 0.178618 (rank : 31) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
RMP_HUMAN
|
||||||
| θ value | 0.21417 (rank : 37) | NC score | 0.077682 (rank : 80) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94763 | Gene names | RMP, C19orf2 | |||
|
Domain Architecture |
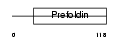
|
|||||
| Description | RNA polymerase II subunit 5-mediating protein (RPB5-mediating protein). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.279714 (rank : 38) | NC score | 0.042313 (rank : 100) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
EGR1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 39) | NC score | 0.008846 (rank : 176) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18146 | Gene names | EGR1, ZNF225 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A) (Transcription factor ETR103) (Zinc finger protein 225) (AT225). | |||||
|
EWS_HUMAN
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.057063 (rank : 88) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
EWS_MOUSE
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.056661 (rank : 89) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
HEN1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.182437 (rank : 28) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02575 | Gene names | NHLH1, HEN1 | |||
|
Domain Architecture |
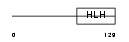
|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
HEN1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 43) | NC score | 0.185849 (rank : 24) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02576 | Gene names | Nhlh1, Hen1 | |||
|
Domain Architecture |
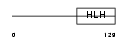
|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
MYCN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 44) | NC score | 0.085149 (rank : 73) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYOG_HUMAN
|
||||||
| θ value | 0.279714 (rank : 45) | NC score | 0.194123 (rank : 21) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P15173 | Gene names | MYOG, MYF4 | |||
|
Domain Architecture |
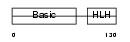
|
|||||
| Description | Myogenin (Myogenic factor 4) (Myf-4). | |||||
|
MYOG_MOUSE
|
||||||
| θ value | 0.279714 (rank : 46) | NC score | 0.188108 (rank : 23) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P12979 | Gene names | Myog | |||
|
Domain Architecture |
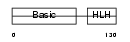
|
|||||
| Description | Myogenin (MYOD1-related protein). | |||||
|
TAL1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 47) | NC score | 0.169327 (rank : 35) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P17542 | Gene names | TAL1, SCL, TCL5 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein (TAL-1 protein) (Stem cell protein) (T-cell leukemia/lymphoma-5 protein). | |||||
|
TAL1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 48) | NC score | 0.169994 (rank : 32) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P22091 | Gene names | Tal1, Scl, Tal-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein homolog (TAL-1 protein) (Stem cell protein). | |||||
|
TB182_HUMAN
|
||||||
| θ value | 0.279714 (rank : 49) | NC score | 0.079922 (rank : 78) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
TTLL4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 50) | NC score | 0.035418 (rank : 111) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14679, Q8WW29 | Gene names | TTLL4, KIAA0173 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
ZN750_HUMAN
|
||||||
| θ value | 0.279714 (rank : 51) | NC score | 0.050938 (rank : 92) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q32MQ0, Q9H899 | Gene names | ZNF750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
CE152_HUMAN
|
||||||
| θ value | 0.365318 (rank : 52) | NC score | 0.026610 (rank : 124) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.365318 (rank : 53) | NC score | 0.071424 (rank : 84) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
NDF4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 54) | NC score | 0.184329 (rank : 27) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9HD90 | Gene names | NEUROD4 | |||
|
Domain Architecture |
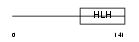
|
|||||
| Description | Neurogenic differentiation factor 4 (NeuroD4). | |||||
|
NDF4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 55) | NC score | 0.181660 (rank : 29) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O09105 | Gene names | Neurod4, Ath3, Atoh3 | |||
|
Domain Architecture |
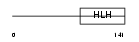
|
|||||
| Description | Neurogenic differentiation factor 4 (NeuroD4) (Atonal protein homolog 3) (Helix-loop-helix protein mATH-3) (MATH3). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.365318 (rank : 56) | NC score | 0.039189 (rank : 104) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.47712 (rank : 57) | NC score | 0.041595 (rank : 101) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
BCAR1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 58) | NC score | 0.034190 (rank : 112) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
LYL1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 59) | NC score | 0.169019 (rank : 36) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P12980, O76102 | Gene names | LYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
MYCN_MOUSE
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.078589 (rank : 79) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 0.813845 (rank : 61) | NC score | 0.022048 (rank : 130) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
DDX5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 62) | NC score | 0.009843 (rank : 175) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.023223 (rank : 128) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
FGD5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.017343 (rank : 152) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
LATS2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.003854 (rank : 189) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TSJ6, Q8CDJ4, Q9JMI3 | Gene names | Lats2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS2 (EC 2.7.11.1) (Large tumor suppressor homolog 2) (Serine/threonine-protein kinase kpm) (Kinase phosphorylated during mitosis protein). | |||||
|
LYL1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.162261 (rank : 40) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P27792 | Gene names | Lyl1, Lyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
STK6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.003277 (rank : 190) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14965, O60445, O75873, Q9BQD6, Q9UPG5 | Gene names | STK6, AIK, ARK1, AURA, BTAK, STK15 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 6 (EC 2.7.11.1) (Serine/threonine kinase 15) (Aurora/IPL1-related kinase 1) (Aurora-related kinase 1) (hARK1) (Aurora-A) (Breast-tumor-amplified kinase). | |||||
|
AKAP8_MOUSE
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.029821 (rank : 120) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBR0, Q9R0L8 | Gene names | Akap8, Akap95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.019890 (rank : 140) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.005235 (rank : 187) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
FIGLA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.164770 (rank : 39) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6QHK4 | Gene names | FIGLA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
IRS1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.035845 (rank : 110) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
K1C9_HUMAN
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.008362 (rank : 177) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35527, O00109, Q14665 | Gene names | KRT9 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 9 (Cytokeratin-9) (CK-9) (Keratin-9) (K9). | |||||
|
MUSC_HUMAN
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.132522 (rank : 44) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O60682, O75946, Q9BRE7 | Gene names | MSC, ABF1 | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Activated B-cell factor 1) (ABF-1). | |||||
|
MUSC_MOUSE
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.133728 (rank : 43) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O88940 | Gene names | Msc, Myor | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Myogenic repressor). | |||||
|
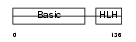
MYF6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.166093 (rank : 38) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P23409, Q53X80, Q6FHI9 | Gene names | MYF6 | |||
|
Domain Architecture |
|
|||||
| Description | Myogenic factor 6 (Myf-6). | |||||
|
MYF6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.166578 (rank : 37) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P15375 | Gene names | Myf6, Myf-6 | |||
|
Domain Architecture |
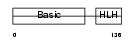
|
|||||
| Description | Myogenic factor 6 (Myf-6) (Herculin). | |||||
|
PRDM1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 78) | NC score | 0.000830 (rank : 193) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 766 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60636 | Gene names | Prdm1, Blimp1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 1 (PR domain-containing protein 1) (Beta-interferon gene positive-regulatory domain I-binding factor) (BLIMP-1). | |||||
|
Z3H7B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 79) | NC score | 0.019172 (rank : 145) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGR2, Q5TFX9, Q8TBT9, Q9H8B6, Q9UGQ9, Q9UGR0, Q9UGR1, Q9UK03, Q9UPW9 | Gene names | ZC3H7B, KIAA1031 | |||
|
Domain Architecture |
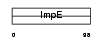
|
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7B (Rotavirus 'X'- associated non-structural protein) (RoXaN). | |||||
|
CDCP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | 0.027354 (rank : 123) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H5V8, Q49UB4, Q6NT71, Q6U9Y2, Q8WU91, Q96QU7, Q9H676, Q9H8C2 | Gene names | CDCP1, TRASK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB domain-containing protein 1 precursor (Transmembrane and associated with src kinases) (Membrane glycoprotein gp140) (Subtractive immunization M plus HEp3-associated 135 kDa protein) (SIMA135) (CD318 antigen). | |||||
|
DAB2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.038869 (rank : 105) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
DAB2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 82) | NC score | 0.036613 (rank : 109) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P98078, Q3U3K1, Q91W56, Q923E1 | Gene names | Dab2, Doc2 | |||
|
Domain Architecture |
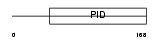
|
|||||
| Description | Disabled homolog 2 (DOC-2) (Mitogen-responsive phosphoprotein). | |||||
|
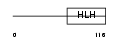
HAND2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 83) | NC score | 0.158677 (rank : 41) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P61296, O95300, O95301, P97833 | Gene names | HAND2, DHAND | |||
|
Domain Architecture |
|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND). | |||||
|
HAND2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 84) | NC score | 0.158677 (rank : 42) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61039, Q61100 | Gene names | Hand2, Dhand, Hed, Thing2 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND) (Helix-loop-helix transcription factor expressed in embryo and deciduum) (Thing-2). | |||||
|
RNPS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.042641 (rank : 98) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.042641 (rank : 99) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
CO4A2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.011938 (rank : 169) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
INVS_HUMAN
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.019905 (rank : 139) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
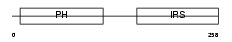
IRS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.048921 (rank : 93) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35568 | Gene names | IRS1 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
K0423_MOUSE
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.033541 (rank : 114) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6A070 | Gene names | Kiaa0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.037836 (rank : 107) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LPHN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.005705 (rank : 183) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94910, Q96IE7, Q9BU07, Q9HAR3 | Gene names | LPHN1, KIAA0821, LEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-1 precursor (Calcium-independent alpha-latrotoxin receptor 1) (Lectomedin-2). | |||||
|
POT15_HUMAN
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.019652 (rank : 142) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.020250 (rank : 135) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.020675 (rank : 133) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 96) | NC score | 0.067045 (rank : 87) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
UTY_HUMAN
|
||||||
| θ value | 2.36792 (rank : 97) | NC score | 0.026117 (rank : 125) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14607, O14608 | Gene names | UTY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed Y chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the Y chromosome). | |||||
|
BHLH4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.119034 (rank : 48) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BGW3, Q71MB7 | Gene names | Bhlhb4, H20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 4 (bHLHB4). | |||||
|
DDX5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.008270 (rank : 178) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
ERC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.013635 (rank : 164) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ERC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.013752 (rank : 162) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.045065 (rank : 97) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
MEFV_HUMAN
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.005385 (rank : 186) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.033284 (rank : 115) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
PRRT1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 105) | NC score | 0.030556 (rank : 119) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35449 | Gene names | Prrt1, Ng5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 1. | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.020733 (rank : 131) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
SH2D3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.016557 (rank : 155) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N5H7, Q5HYE5, Q6UY42, Q8N6X3, Q9Y2X5 | Gene names | SH2D3C, NSP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (Novel SH2-containing protein 3). | |||||
|
SNCAP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.020139 (rank : 137) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6H5 | Gene names | SNCAIP | |||
|
Domain Architecture |

|
|||||
| Description | Synphilin-1 (Alpha-synuclein-interacting protein). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.034041 (rank : 113) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.013884 (rank : 160) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
BTBDC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.028922 (rank : 121) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
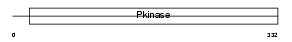
CDK8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.001865 (rank : 191) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49336 | Gene names | CDK8 | |||
|
Domain Architecture |
|
|||||
| Description | Cell division protein kinase 8 (EC 2.7.11.22) (EC 2.7.11.23) (Protein kinase K35). | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.011606 (rank : 171) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.013843 (rank : 161) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
DJBP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.017308 (rank : 154) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P1E8, Q5DTV8, Q6PGK8, Q9D2D8 | Gene names | Djbp, Kiaa1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein. | |||||
|
FBX24_MOUSE
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.040920 (rank : 102) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D417 | Gene names | Fbxo24, Fbx24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 24. | |||||
|
K1802_HUMAN
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.007538 (rank : 181) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MITF_MOUSE
|
||||||
| θ value | 4.03905 (rank : 118) | NC score | 0.045484 (rank : 96) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.018022 (rank : 150) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
SOX8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.005529 (rank : 185) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q04886 | Gene names | Sox8, Sox-8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
BHLH4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.107052 (rank : 55) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NDY6 | Gene names | BHLHB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 4 (bHLHB4). | |||||
|
CDSN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.047615 (rank : 95) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15517, O43509, Q5SQ85, Q5STD2, Q7LA70, Q7LA71, Q86Z04, Q8IZU4, Q8IZU5, Q8IZU6, Q8N5P3, Q95IF9, Q9NP52, Q9NPE0, Q9NPG5, Q9NRH4, Q9NRH5, Q9NRH6, Q9NRH7, Q9NRH8, Q9UBH8, Q9UIN6, Q9UIN7, Q9UIN8, Q9UIN9, Q9UIP0 | Gene names | CDSN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor (S protein). | |||||
|
CT175_HUMAN
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.019325 (rank : 144) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96MK2, Q5QPB6, Q9NQQ2 | Gene names | C20orf175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf175. | |||||
|
DMXL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.010890 (rank : 173) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PNC0, Q3TLG8, Q6PHM6, Q8BQF2 | Gene names | Dmxl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
FA21C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.019636 (rank : 143) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.013696 (rank : 163) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.025108 (rank : 127) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
MCCA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.012165 (rank : 168) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96RQ3, Q59ES4, Q9H959, Q9NS97 | Gene names | MCCC1, MCCA | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
MITF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.040632 (rank : 103) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
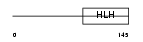
OLIG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.095532 (rank : 64) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8TAK6, Q7RTS0 | Gene names | OLIG1 | |||
|
Domain Architecture |
|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1). | |||||
|
SAKS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.020092 (rank : 138) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q04323, Q9BV93, Q9BVV5 | Gene names | SAKS1 | |||
|
Domain Architecture |

|
|||||
| Description | SAPK substrate protein 1 (UBA/UBX 33.3 kDa protein). | |||||
|
TFEB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.033179 (rank : 116) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.031232 (rank : 117) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.038583 (rank : 106) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
BRD7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.018993 (rank : 147) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NPI1, Q4VC09, Q8N2L9, Q96KA4, Q9BV48, Q9UH59 | Gene names | BRD7, BP75, CELTIX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein) (Protein CELTIX-1). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.014949 (rank : 158) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CD248_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.009997 (rank : 174) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CROP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.055762 (rank : 90) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95232, Q6PHR9, Q9NUY0, Q9P2S7 | Gene names | CROP, CREAP1, O48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein (cAMP regulatory element-associated protein 1) (CRE-associated protein 1) (CREAP-1) (Luc7A) (Okadaic acid-inducible phosphoprotein OA48-18). | |||||
|
CROP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.055463 (rank : 91) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5SUF2, Q3U9D5, Q8BUJ5, Q921Z3, Q9CRS7, Q9CTY4 | Gene names | Crop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein. | |||||
|
DACT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.014451 (rank : 159) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYF0, Q86TY0 | Gene names | DACT1, DPR1, HNG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (hDPR1) (Heptacellular carcinoma novel gene 3 protein). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.048197 (rank : 94) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
F125A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.018379 (rank : 148) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q78HU3 | Gene names | Fam125a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM125A. | |||||
|
HNF1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.018072 (rank : 149) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35680 | Gene names | TCF2, HNF1B | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-beta (HNF-1beta) (HNF-1B) (Variant hepatic nuclear factor 1) (VHNF1) (Homeoprotein LFB3) (Transcription factor 2) (TCF-2). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.013333 (rank : 165) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
IPP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.020575 (rank : 134) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DCL8, Q3UD25, Q8C1A6 | Gene names | Ppp1r2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase inhibitor 2 (IPP-2). | |||||
|
PER2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.020713 (rank : 132) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
REN3B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.025850 (rank : 126) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BZI7, Q9H1J0 | Gene names | UPF3B, RENT3B, UPF3X | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 3B (Nonsense mRNA reducing factor 3B) (Up-frameshift suppressor 3 homolog B) (hUpf3B) (hUpf3p-X). | |||||
|
SMAD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.005601 (rank : 184) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70340, P70442, Q9CYK6 | Gene names | Smad1, Madh1, Madr1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Mothers-against-DPP-related 1) (mMad1) (Dwarfin-A) (Dwf-A). | |||||
|
SRP72_HUMAN
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.031219 (rank : 118) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O76094 | Gene names | SRP72 | |||
|
Domain Architecture |

|
|||||
| Description | Signal recognition particle 72 kDa protein (SRP72). | |||||
|
B3A3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.008218 (rank : 179) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16283 | Gene names | Slc4a3, Ae3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3). | |||||
|
BRE1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.017784 (rank : 151) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
BRE1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.019143 (rank : 146) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
CAC1F_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.004189 (rank : 188) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CJ006_MOUSE
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.015698 (rank : 156) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P9P0, Q8C041, Q8C0J5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf6 homolog. | |||||
|
DIAP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.011724 (rank : 170) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
ENAH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.011389 (rank : 172) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.005761 (rank : 182) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.019708 (rank : 141) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
K0423_HUMAN
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.017341 (rank : 153) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.022220 (rank : 129) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NOTC3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.000170 (rank : 194) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
PCD17_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | -0.000258 (rank : 195) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14917 | Gene names | PCDH17, PCDH68, PCH68 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-17 precursor (Protocadherin-68). | |||||
|
PRRT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.020224 (rank : 136) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99946, Q96DW3, Q96NQ8 | Gene names | PRRT1, C6orf31, NG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 1. | |||||
|
PTF1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.126338 (rank : 45) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7RTS3, Q9HC25 | Gene names | PTF1A, PTF1P48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
PTF1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.125853 (rank : 46) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9QX98, Q9QYF5, Q9QYF6 | Gene names | Ptf1a, Ptf1p48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
PTN14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.001265 (rank : 192) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62130 | Gene names | Ptpn14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP36). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.013330 (rank : 166) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
T240L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.007623 (rank : 180) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q71F56, Q68DN4, Q9H8C0, Q9NSY9, Q9UFD8, Q9UPX5 | Gene names | THRAP2, KIAA1025, PROSIT240, TRAP240L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
ASCL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.080616 (rank : 76) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P50553, Q9BQ30 | Gene names | ASCL1, ASH1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (HASH1). | |||||
|
ASCL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.081248 (rank : 74) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q02067 | Gene names | Ascl1, Ash1, Mash-1, Mash1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (Mash-1). | |||||
|
ASCL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.098348 (rank : 60) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99929, Q9UM68 | Gene names | ASCL2 | |||
|
Domain Architecture |
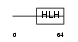
|
|||||
| Description | Achaete-scute homolog 2 (Mash2) (HASH2). | |||||
|
ASCL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.081176 (rank : 75) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O35885, Q9WUJ7 | Gene names | Ascl2, Mash2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash-2). | |||||
|
ASCL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.090963 (rank : 68) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NQ33, Q8WYQ6 | Gene names | ASCL3, HASH3, SGN1 | |||
|
Domain Architecture |
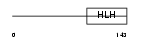
|
|||||
| Description | Achaete-scute homolog 3 (bHLH transcriptional regulator Sgn-1). | |||||
|
ASCL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.093806 (rank : 65) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JJR7 | Gene names | Ascl3, Mash3, Sgn1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 3 (bHLH transcriptional regulator Sgn-1) (Mash- 3). | |||||
|
HAND1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.095776 (rank : 63) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O96004 | Gene names | HAND1, EHAND | |||
|
Domain Architecture |
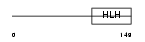
|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND). | |||||
|
HAND1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.096632 (rank : 62) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q64279, Q61099 | Gene names | Hand1, Ehand, Hxt, Thing1 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND) (Helix-loop-helix transcription factor expressed in extraembryonic mesoderm and trophoblast) (Thing-1) (Th1). | |||||
|
NGN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.107914 (rank : 53) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92886, Q96HE1 | Gene names | NEUROG1, NEUROD3, NGN, NGN1 | |||
|
Domain Architecture |
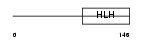
|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein). | |||||
|
NGN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.110754 (rank : 50) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P70660 | Gene names | Neurog1, Ath4c, Neurod3, Ngn, Ngn1 | |||
|
Domain Architecture |
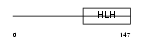
|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein) (Helix-loop-helix protein mATH-4C) (MATH4C). | |||||
|
NGN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.104496 (rank : 58) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9H2A3, Q8N416 | Gene names | NEUROG2, NGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogenin 2. | |||||
|
NGN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.104452 (rank : 59) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P70447, P70237 | Gene names | Neurog2, Ath4a, Atoh4, Ngn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 2 (Atonal protein homolog 4) (Helix-loop-helix protein mATH-4A) (MATH4A). | |||||
|
NGN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.105210 (rank : 57) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y4Z2, Q9BY24 | Gene names | NEUROG3, NGN3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3. | |||||
|
NGN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.106146 (rank : 56) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P70661 | Gene names | Neurog3, Ath4b, Atoh5, Ngn3 | |||
|
Domain Architecture |
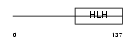
|
|||||
| Description | Neurogenin 3 (Atonal protein homolog 5) (Helix-loop-helix protein mATH-4B) (MATH4B). | |||||
|
OLIG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.071682 (rank : 83) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JKN5 | Gene names | Olig1 | |||
|
Domain Architecture |
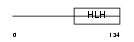
|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1) (Oligodendrocyte- specific bHLH transcription factor 1). | |||||
|
OLIG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.087572 (rank : 72) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13516, Q86X04, Q9NZ14 | Gene names | OLIG2, BHLHB1, PRKCBP2, RACK17 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2) (Basic helix-loop- helix protein class B 1) (Protein kinase C-binding protein RACK17) (Protein kinase C-binding protein 2). | |||||
|
OLIG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.089663 (rank : 69) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9EQW6, Q9JKN4 | Gene names | Olig2 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2). | |||||
|
OLIG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.069082 (rank : 86) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7RTU3, Q8N8Q0 | Gene names | OLIG3, BHLHB7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oligodendrocyte transcription factor 3 (Oligo3) (Basic helix-loop- helix domain-containing class B protein 7). | |||||
|
OLIG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.070750 (rank : 85) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6PFG8, Q9EQW5 | Gene names | Olig3, Bhlhb7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oligodendrocyte transcription factor 3 (Oligo3) (Oligodendrocyte- specific bHLH transcription factor 3) (Basic helix-loop-helix domain- containing class B protein 7). | |||||
|
SCX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.108802 (rank : 52) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q64124 | Gene names | Scx | |||
|
Domain Architecture |
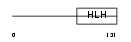
|
|||||
| Description | Basic helix-loop-helix transcription factor scleraxis. | |||||
|
TAL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.107911 (rank : 54) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q16559 | Gene names | TAL2 | |||
|
Domain Architecture |
|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein (TAL-2 protein). | |||||
|
TAL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.109338 (rank : 51) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q62282 | Gene names | Tal2 | |||
|
Domain Architecture |
|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein homolog (TAL-2 protein). | |||||
|
TCF15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.093705 (rank : 66) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q12870, Q9NQQ1 | Gene names | TCF15, BHLHEC2 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis). | |||||
|
TCF15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.097677 (rank : 61) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q60756, Q60788 | Gene names | Tcf15, Bhlhec2 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis) (Meso1). | |||||
|
TCF21_HUMAN
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.088645 (rank : 70) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43680, O43545, Q9BZ14 | Gene names | TCF21, POD1 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor 21 (Podocyte-expressed 1) (Pod-1) (Epicardin) (Capsulin). | |||||
|
TCF21_MOUSE
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.088213 (rank : 71) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O35437, Q3U023 | Gene names | Tcf21, Pod1 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor 21 (Podocyte-expressed 1) (Pod-1) (Epicardin) (Capsulin). | |||||
|
TFAP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.075835 (rank : 81) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
ITF2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 168 | |
| SwissProt Accessions | P15884, Q15439, Q15440 | Gene names | TCF4, ITF2, SEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (SL3-3 enhancer factor 2) (SEF-2). | |||||
|
ITF2_MOUSE
|
||||||
| NC score | 0.991501 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 130 | |
| SwissProt Accessions | Q60722, Q60721, Q62211, Q64072, Q80UE8 | Gene names | Tcf4, Itf2, Sef2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (MITF-2) (SL3-3 enhancer factor 2) (SEF-2) (Class A helix-loop-helix transcription factor ME2). | |||||
|
HTF4_MOUSE
|
||||||
| NC score | 0.964384 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q61286 | Gene names | Tcf12, Alf1, Meb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4) (Class A helix-loop-helix transcription factor ME1). | |||||
|
HTF4_HUMAN
|
||||||
| NC score | 0.963098 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q99081 | Gene names | TCF12, HEB, HTF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4). | |||||
|
TFE2_MOUSE
|
||||||
| NC score | 0.946347 (rank : 5) | θ value | 4.15819e-130 (rank : 6) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P15806, Q3U153, Q8CAH9, Q8VCY4, Q922S2, Q99MB8, Q9CYJ4 | Gene names | Tcf3, Alf2, Me2, Tcfe2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Transcription factor A1). | |||||
|
TFE2_HUMAN
|
||||||
| NC score | 0.946189 (rank : 6) | θ value | 3.06949e-141 (rank : 5) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P15923, P15883, Q14208, Q14635, Q14636, Q9UPI9 | Gene names | TCF3, E2A, ITF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Immunoglobulin transcription factor 1) (Transcription factor ITF-1) (Kappa-E2-binding factor). | |||||
|
ATOH1_MOUSE
|
||||||
| NC score | 0.240755 (rank : 7) | θ value | 0.00509761 (rank : 10) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P48985 | Gene names | Atoh1, Ath1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein mATH-1) (MATH1). | |||||
|
MYF5_MOUSE
|
||||||
| NC score | 0.226738 (rank : 8) | θ value | 0.0563607 (rank : 21) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P24699, Q543W7 | Gene names | Myf5, Myf-5 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
FIGLA_MOUSE
|
||||||
| NC score | 0.224488 (rank : 9) | θ value | 0.0736092 (rank : 23) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O55208 | Gene names | Figla | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
MYF5_HUMAN
|
||||||
| NC score | 0.223829 (rank : 10) | θ value | 0.0563607 (rank : 20) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P13349 | Gene names | MYF5 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
ATOH1_HUMAN
|
||||||
| NC score | 0.216088 (rank : 11) | θ value | 0.0148317 (rank : 14) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92858 | Gene names | ATOH1, ATH1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein hATH-1). | |||||
|
BHLH8_MOUSE
|
||||||
| NC score | 0.212558 (rank : 12) | θ value | 0.00869519 (rank : 11) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
MYOD1_MOUSE
|
||||||
| NC score | 0.207464 (rank : 13) | θ value | 0.0961366 (rank : 27) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P10085 | Gene names | Myod1, Myod | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1. | |||||
|
MYOD1_HUMAN
|
||||||
| NC score | 0.205454 (rank : 14) | θ value | 0.0961366 (rank : 26) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P15172 | Gene names | MYOD1, MYF3, MYOD | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1 (Myogenic factor 3) (Myf-3). | |||||
|
TWST1_HUMAN
|
||||||
| NC score | 0.199023 (rank : 15) | θ value | 0.0736092 (rank : 24) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15672, Q92487, Q99804 | Gene names | TWIST1, TWIST | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (H-twist). | |||||
|
TWST1_MOUSE
|
||||||
| NC score | 0.198711 (rank : 16) | θ value | 0.0736092 (rank : 25) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P26687 | Gene names | Twist1, Twist | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (M-twist). | |||||
|
TWST2_MOUSE
|
||||||
| NC score | 0.196661 (rank : 17) | θ value | 0.0961366 (rank : 31) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9D030 | Gene names | Twist2, Dermo1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST2_HUMAN
|
||||||
| NC score | 0.196458 (rank : 18) | θ value | 0.0961366 (rank : 30) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8WVJ9 | Gene names | TWIST2, DERMO1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
NDF2_HUMAN
|
||||||
| NC score | 0.194944 (rank : 19) | θ value | 0.0431538 (rank : 16) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q15784, Q8TBI7, Q9UQC6 | Gene names | NEUROD2, NDRF | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
NDF2_MOUSE
|
||||||
| NC score | 0.194568 (rank : 20) | θ value | 0.0431538 (rank : 17) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62414, Q61952, Q925V5 | Gene names | Neurod2, Ndrf | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
MYOG_HUMAN
|
||||||
| NC score | 0.194123 (rank : 21) | θ value | 0.279714 (rank : 45) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P15173 | Gene names | MYOG, MYF4 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenin (Myogenic factor 4) (Myf-4). | |||||
|
BHLH8_HUMAN
|
||||||
| NC score | 0.190246 (rank : 22) | θ value | 0.0736092 (rank : 22) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q7RTS1 | Gene names | BHLHB8, MIST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
MYOG_MOUSE
|
||||||
| NC score | 0.188108 (rank : 23) | θ value | 0.279714 (rank : 46) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P12979 | Gene names | Myog | |||
|
Domain Architecture |

|
|||||
| Description | Myogenin (MYOD1-related protein). | |||||
|
HEN1_MOUSE
|
||||||
| NC score | 0.185849 (rank : 24) | θ value | 0.279714 (rank : 43) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02576 | Gene names | Nhlh1, Hen1 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
HEN2_HUMAN
|
||||||
| NC score | 0.185513 (rank : 25) | θ value | 0.125558 (rank : 32) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02577 | Gene names | NHLH2, HEN2 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
HEN2_MOUSE
|
||||||
| NC score | 0.185398 (rank : 26) | θ value | 0.125558 (rank : 33) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q64221 | Gene names | Nhlh2, Hen2 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
NDF4_HUMAN
|
||||||
| NC score | 0.184329 (rank : 27) | θ value | 0.365318 (rank : 54) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9HD90 | Gene names | NEUROD4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 4 (NeuroD4). | |||||
|
HEN1_HUMAN
|
||||||
| NC score | 0.182437 (rank : 28) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02575 | Gene names | NHLH1, HEN1 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
NDF4_MOUSE
|
||||||
| NC score | 0.181660 (rank : 29) | θ value | 0.365318 (rank : 55) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O09105 | Gene names | Neurod4, Ath3, Atoh3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 4 (NeuroD4) (Atonal protein homolog 3) (Helix-loop-helix protein mATH-3) (MATH3). | |||||
|
NDF1_HUMAN
|
||||||
| NC score | 0.178979 (rank : 30) | θ value | 0.125558 (rank : 34) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
NDF1_MOUSE
|
||||||
| NC score | 0.178618 (rank : 31) | θ value | 0.163984 (rank : 36) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
TAL1_MOUSE
|
||||||
| NC score | 0.169994 (rank : 32) | θ value | 0.279714 (rank : 48) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P22091 | Gene names | Tal1, Scl, Tal-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein homolog (TAL-1 protein) (Stem cell protein). | |||||
|
NDF6_MOUSE
|
||||||
| NC score | 0.169849 (rank : 33) | θ value | 0.0961366 (rank : 29) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P48986 | Gene names | Neurod6, Ath2, Atoh2, Nex1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 6 (NeuroD6) (Atonal protein homolog 2) (Helix-loop-helix protein mATH-2) (MATH2) (NEX-1 protein). | |||||
|
NDF6_HUMAN
|
||||||
| NC score | 0.169788 (rank : 34) | θ value | 0.0961366 (rank : 28) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96NK8, Q9H3H6 | Gene names | NEUROD6 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 6 (NeuroD6). | |||||
|
TAL1_HUMAN
|
||||||
| NC score | 0.169327 (rank : 35) | θ value | 0.279714 (rank : 47) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P17542 | Gene names | TAL1, SCL, TCL5 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein (TAL-1 protein) (Stem cell protein) (T-cell leukemia/lymphoma-5 protein). | |||||
|
LYL1_HUMAN
|
||||||
| NC score | 0.169019 (rank : 36) | θ value | 0.62314 (rank : 59) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P12980, O76102 | Gene names | LYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
MYF6_MOUSE
|
||||||
| NC score | 0.166578 (rank : 37) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P15375 | Gene names | Myf6, Myf-6 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 6 (Myf-6) (Herculin). | |||||
|
MYF6_HUMAN
|
||||||
| NC score | 0.166093 (rank : 38) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P23409, Q53X80, Q6FHI9 | Gene names | MYF6 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 6 (Myf-6). | |||||
|
FIGLA_HUMAN
|
||||||
| NC score | 0.164770 (rank : 39) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6QHK4 | Gene names | FIGLA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
LYL1_MOUSE
|
||||||
| NC score | 0.162261 (rank : 40) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P27792 | Gene names | Lyl1, Lyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
HAND2_HUMAN
|
||||||
| NC score | 0.158677 (rank : 41) | θ value | 1.81305 (rank : 83) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P61296, O95300, O95301, P97833 | Gene names | HAND2, DHAND | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND). | |||||
|
HAND2_MOUSE
|
||||||
| NC score | 0.158677 (rank : 42) | θ value | 1.81305 (rank : 84) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61039, Q61100 | Gene names | Hand2, Dhand, Hed, Thing2 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND) (Helix-loop-helix transcription factor expressed in embryo and deciduum) (Thing-2). | |||||
|
MUSC_MOUSE
|
||||||
| NC score | 0.133728 (rank : 43) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O88940 | Gene names | Msc, Myor | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Myogenic repressor). | |||||
|
MUSC_HUMAN
|
||||||
| NC score | 0.132522 (rank : 44) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O60682, O75946, Q9BRE7 | Gene names | MSC, ABF1 | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Activated B-cell factor 1) (ABF-1). | |||||
|
PTF1A_HUMAN
|
||||||
| NC score | 0.126338 (rank : 45) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7RTS3, Q9HC25 | Gene names | PTF1A, PTF1P48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
PTF1A_MOUSE
|
||||||
| NC score | 0.125853 (rank : 46) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9QX98, Q9QYF5, Q9QYF6 | Gene names | Ptf1a, Ptf1p48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
HORN_HUMAN
|
||||||
| NC score | 0.125766 (rank : 47) | θ value | 0.000602161 (rank : 9) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
BHLH4_MOUSE
|
||||||
| NC score | 0.119034 (rank : 48) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BGW3, Q71MB7 | Gene names | Bhlhb4, H20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 4 (bHLHB4). | |||||
|
HORN_MOUSE
|
||||||
| NC score | 0.117388 (rank : 49) | θ value | 7.1131e-05 (rank : 7) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
NGN1_MOUSE
|
||||||
| NC score | 0.110754 (rank : 50) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P70660 | Gene names | Neurog1, Ath4c, Neurod3, Ngn, Ngn1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein) (Helix-loop-helix protein mATH-4C) (MATH4C). | |||||
|
TAL2_MOUSE
|
||||||
| NC score | 0.109338 (rank : 51) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q62282 | Gene names | Tal2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein homolog (TAL-2 protein). | |||||
|
SCX_MOUSE
|
||||||
| NC score | 0.108802 (rank : 52) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q64124 | Gene names | Scx | |||
|
Domain Architecture |

|
|||||
| Description | Basic helix-loop-helix transcription factor scleraxis. | |||||
|
NGN1_HUMAN
|
||||||
| NC score | 0.107914 (rank : 53) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92886, Q96HE1 | Gene names | NEUROG1, NEUROD3, NGN, NGN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein). | |||||
|
TAL2_HUMAN
|
||||||
| NC score | 0.107911 (rank : 54) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q16559 | Gene names | TAL2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein (TAL-2 protein). | |||||
|
BHLH4_HUMAN
|
||||||
| NC score | 0.107052 (rank : 55) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NDY6 | Gene names | BHLHB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 4 (bHLHB4). | |||||
|
NGN3_MOUSE
|
||||||
| NC score | 0.106146 (rank : 56) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P70661 | Gene names | Neurog3, Ath4b, Atoh5, Ngn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3 (Atonal protein homolog 5) (Helix-loop-helix protein mATH-4B) (MATH4B). | |||||
|
NGN3_HUMAN
|
||||||
| NC score | 0.105210 (rank : 57) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y4Z2, Q9BY24 | Gene names | NEUROG3, NGN3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3. | |||||
|
NGN2_HUMAN
|
||||||
| NC score | 0.104496 (rank : 58) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9H2A3, Q8N416 | Gene names | NEUROG2, NGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogenin 2. | |||||
|
NGN2_MOUSE
|
||||||
| NC score | 0.104452 (rank : 59) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P70447, P70237 | Gene names | Neurog2, Ath4a, Atoh4, Ngn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 2 (Atonal protein homolog 4) (Helix-loop-helix protein mATH-4A) (MATH4A). | |||||
|
ASCL2_HUMAN
|
||||||
| NC score | 0.098348 (rank : 60) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99929, Q9UM68 | Gene names | ASCL2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash2) (HASH2). | |||||
|
TCF15_MOUSE
|
||||||
| NC score | 0.097677 (rank : 61) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q60756, Q60788 | Gene names | Tcf15, Bhlhec2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis) (Meso1). | |||||
|
HAND1_MOUSE
|
||||||
| NC score | 0.096632 (rank : 62) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q64279, Q61099 | Gene names | Hand1, Ehand, Hxt, Thing1 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND) (Helix-loop-helix transcription factor expressed in extraembryonic mesoderm and trophoblast) (Thing-1) (Th1). | |||||
|
HAND1_HUMAN
|
||||||
| NC score | 0.095776 (rank : 63) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O96004 | Gene names | HAND1, EHAND | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND). | |||||
|
OLIG1_HUMAN
|
||||||
| NC score | 0.095532 (rank : 64) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8TAK6, Q7RTS0 | Gene names | OLIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1). | |||||
|
ASCL3_MOUSE
|
||||||
| NC score | 0.093806 (rank : 65) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JJR7 | Gene names | Ascl3, Mash3, Sgn1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 3 (bHLH transcriptional regulator Sgn-1) (Mash- 3). | |||||
|
TCF15_HUMAN
|
||||||
| NC score | 0.093705 (rank : 66) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q12870, Q9NQQ1 | Gene names | TCF15, BHLHEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.093258 (rank : 67) | θ value | 0.00035302 (rank : 8) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ASCL3_HUMAN
|
||||||
| NC score | 0.090963 (rank : 68) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NQ33, Q8WYQ6 | Gene names | ASCL3, HASH3, SGN1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 3 (bHLH transcriptional regulator Sgn-1). | |||||
|
OLIG2_MOUSE
|
||||||
| NC score | 0.089663 (rank : 69) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9EQW6, Q9JKN4 | Gene names | Olig2 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2). | |||||
|
TCF21_HUMAN
|
||||||
| NC score | 0.088645 (rank : 70) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43680, O43545, Q9BZ14 | Gene names | TCF21, POD1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 21 (Podocyte-expressed 1) (Pod-1) (Epicardin) (Capsulin). | |||||
|
TCF21_MOUSE
|
||||||
| NC score | 0.088213 (rank : 71) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O35437, Q3U023 | Gene names | Tcf21, Pod1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 21 (Podocyte-expressed 1) (Pod-1) (Epicardin) (Capsulin). | |||||
|
OLIG2_HUMAN
|
||||||
| NC score | 0.087572 (rank : 72) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13516, Q86X04, Q9NZ14 | Gene names | OLIG2, BHLHB1, PRKCBP2, RACK17 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2) (Basic helix-loop- helix protein class B 1) (Protein kinase C-binding protein RACK17) (Protein kinase C-binding protein 2). | |||||
|
MYCN_HUMAN
|
||||||
| NC score | 0.085149 (rank : 73) | θ value | 0.279714 (rank : 44) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
ASCL1_MOUSE
|
||||||
| NC score | 0.081248 (rank : 74) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q02067 | Gene names | Ascl1, Ash1, Mash-1, Mash1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (Mash-1). | |||||
|
ASCL2_MOUSE
|
||||||
| NC score | 0.081176 (rank : 75) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O35885, Q9WUJ7 | Gene names | Ascl2, Mash2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash-2). | |||||
|
ASCL1_HUMAN
|
||||||
| NC score | 0.080616 (rank : 76) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P50553, Q9BQ30 | Gene names | ASCL1, ASH1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (HASH1). | |||||
|
PPRB_MOUSE
|
||||||
| NC score | 0.080152 (rank : 77) | θ value | 0.0113563 (rank : 13) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
TB182_HUMAN
|
||||||
| NC score | 0.079922 (rank : 78) | θ value | 0.279714 (rank : 49) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
MYCN_MOUSE
|
||||||
| NC score | 0.078589 (rank : 79) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
RMP_HUMAN
|
||||||
| NC score | 0.077682 (rank : 80) | θ value | 0.21417 (rank : 37) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94763 | Gene names | RMP, C19orf2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit 5-mediating protein (RPB5-mediating protein). | |||||
|
TFAP4_HUMAN
|
||||||
| NC score | 0.075835 (rank : 81) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.075185 (rank : 82) | θ value | 0.0431538 (rank : 18) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
OLIG1_MOUSE
|
||||||
| NC score | 0.071682 (rank : 83) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JKN5 | Gene names | Olig1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1) (Oligodendrocyte- specific bHLH transcription factor 1). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.071424 (rank : 84) | θ value | 0.365318 (rank : 53) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
OLIG3_MOUSE
|
||||||
| NC score | 0.070750 (rank : 85) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6PFG8, Q9EQW5 | Gene names | Olig3, Bhlhb7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oligodendrocyte transcription factor 3 (Oligo3) (Oligodendrocyte- specific bHLH transcription factor 3) (Basic helix-loop-helix domain- containing class B protein 7). | |||||
|
OLIG3_HUMAN
|
||||||
| NC score | 0.069082 (rank : 86) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7RTU3, Q8N8Q0 | Gene names | OLIG3, BHLHB7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oligodendrocyte transcription factor 3 (Oligo3) (Basic helix-loop- helix domain-containing class B protein 7). | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.067045 (rank : 87) | θ value | 2.36792 (rank : 96) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
EWS_HUMAN
|
||||||
| NC score | 0.057063 (rank : 88) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
EWS_MOUSE
|
||||||
| NC score | 0.056661 (rank : 89) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
CROP_HUMAN
|
||||||
| NC score | 0.055762 (rank : 90) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95232, Q6PHR9, Q9NUY0, Q9P2S7 | Gene names | CROP, CREAP1, O48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein (cAMP regulatory element-associated protein 1) (CRE-associated protein 1) (CREAP-1) (Luc7A) (Okadaic acid-inducible phosphoprotein OA48-18). | |||||
|
CROP_MOUSE
|
||||||
| NC score | 0.055463 (rank : 91) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5SUF2, Q3U9D5, Q8BUJ5, Q921Z3, Q9CRS7, Q9CTY4 | Gene names | Crop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein. | |||||
|
ZN750_HUMAN
|
||||||
| NC score | 0.050938 (rank : 92) | θ value | 0.279714 (rank : 51) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q32MQ0, Q9H899 | Gene names | ZNF750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
IRS1_HUMAN
|
||||||
| NC score | 0.048921 (rank : 93) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35568 | Gene names | IRS1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.048197 (rank : 94) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
CDSN_HUMAN
|
||||||
| NC score | 0.047615 (rank : 95) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15517, O43509, Q5SQ85, Q5STD2, Q7LA70, Q7LA71, Q86Z04, Q8IZU4, Q8IZU5, Q8IZU6, Q8N5P3, Q95IF9, Q9NP52, Q9NPE0, Q9NPG5, Q9NRH4, Q9NRH5, Q9NRH6, Q9NRH7, Q9NRH8, Q9UBH8, Q9UIN6, Q9UIN7, Q9UIN8, Q9UIN9, Q9UIP0 | Gene names | CDSN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor (S protein). | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.045484 (rank : 96) | θ value | 4.03905 (rank : 118) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.045065 (rank : 97) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
RNPS1_HUMAN
|
||||||
| NC score | 0.042641 (rank : 98) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| NC score | 0.042641 (rank : 99) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.042313 (rank : 100) | θ value | 0.279714 (rank : 38) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.041595 (rank : 101) | θ value | 0.47712 (rank : 57) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
FBX24_MOUSE
|
||||||
| NC score | 0.040920 (rank : 102) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D417 | Gene names | Fbxo24, Fbx24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 24. | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.040632 (rank : 103) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.039189 (rank : 104) | θ value | 0.365318 (rank : 56) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
DAB2_HUMAN
|
||||||
| NC score | 0.038869 (rank : 105) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.038583 (rank : 106) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.037836 (rank : 107) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LRRF2_HUMAN
|
||||||
| NC score | 0.037505 (rank : 108) | θ value | 0.0563607 (rank : 19) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
DAB2_MOUSE
|
||||||
| NC score | 0.036613 (rank : 109) | θ value | 1.81305 (rank : 82) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P98078, Q3U3K1, Q91W56, Q923E1 | Gene names | Dab2, Doc2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (DOC-2) (Mitogen-responsive phosphoprotein). | |||||
|
IRS1_MOUSE
|
||||||
| NC score | 0.035845 (rank : 110) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
TTLL4_HUMAN
|
||||||
| NC score | 0.035418 (rank : 111) | θ value | 0.279714 (rank : 50) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14679, Q8WW29 | Gene names | TTLL4, KIAA0173 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
BCAR1_HUMAN
|
||||||
| NC score | 0.034190 (rank : 112) | θ value | 0.62314 (rank : 58) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.034041 (rank : 113) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
K0423_MOUSE
|
||||||
| NC score | 0.033541 (rank : 114) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6A070 | Gene names | Kiaa0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.033284 (rank : 115) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.033179 (rank : 116) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.031232 (rank : 117) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
SRP72_HUMAN
|
||||||
| NC score | 0.031219 (rank : 118) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O76094 | Gene names | SRP72 | |||
|
Domain Architecture |

|
|||||
| Description | Signal recognition particle 72 kDa protein (SRP72). | |||||
|
PRRT1_MOUSE
|
||||||
| NC score | 0.030556 (rank : 119) | θ value | 3.0926 (rank : 105) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35449 | Gene names | Prrt1, Ng5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 1. | |||||
|
AKAP8_MOUSE
|
||||||
| NC score | 0.029821 (rank : 120) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBR0, Q9R0L8 | Gene names | Akap8, Akap95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
BTBDC_HUMAN
|
||||||
| NC score | 0.028922 (rank : 121) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.027476 (rank : 122) | θ value | 0.163984 (rank : 35) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
CDCP1_HUMAN
|
||||||
| NC score | 0.027354 (rank : 123) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H5V8, Q49UB4, Q6NT71, Q6U9Y2, Q8WU91, Q96QU7, Q9H676, Q9H8C2 | Gene names | CDCP1, TRASK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB domain-containing protein 1 precursor (Transmembrane and associated with src kinases) (Membrane glycoprotein gp140) (Subtractive immunization M plus HEp3-associated 135 kDa protein) (SIMA135) (CD318 antigen). | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.026610 (rank : 124) | θ value | 0.365318 (rank : 52) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
UTY_HUMAN
|
||||||
| NC score | 0.026117 (rank : 125) | θ value | 2.36792 (rank : 97) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14607, O14608 | Gene names | UTY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed Y chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the Y chromosome). | |||||
|
REN3B_HUMAN
|
||||||
| NC score | 0.025850 (rank : 126) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BZI7, Q9H1J0 | Gene names | UPF3B, RENT3B, UPF3X | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 3B (Nonsense mRNA reducing factor 3B) (Up-frameshift suppressor 3 homolog B) (hUpf3B) (hUpf3p-X). | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.025108 (rank : 127) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.023223 (rank : 128) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.022220 (rank : 129) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.022048 (rank : 130) | θ value | 0.813845 (rank : 61) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.020733 (rank : 131) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.020713 (rank : 132) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.020675 (rank : 133) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
IPP2_MOUSE
|
||||||
| NC score | 0.020575 (rank : 134) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DCL8, Q3UD25, Q8C1A6 | Gene names | Ppp1r2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase inhibitor 2 (IPP-2). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.020250 (rank : 135) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
PRRT1_HUMAN
|
||||||
| NC score | 0.020224 (rank : 136) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99946, Q96DW3, Q96NQ8 | Gene names | PRRT1, C6orf31, NG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 1. | |||||
|
SNCAP_HUMAN
|
||||||
| NC score | 0.020139 (rank : 137) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6H5 | Gene names | SNCAIP | |||
|
Domain Architecture |

|
|||||
| Description | Synphilin-1 (Alpha-synuclein-interacting protein). | |||||
|
SAKS1_HUMAN
|
||||||
| NC score | 0.020092 (rank : 138) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q04323, Q9BV93, Q9BVV5 | Gene names | SAKS1 | |||
|
Domain Architecture |

|
|||||
| Description | SAPK substrate protein 1 (UBA/UBX 33.3 kDa protein). | |||||
|
INVS_HUMAN
|
||||||
| NC score | 0.019905 (rank : 139) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.019890 (rank : 140) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.019708 (rank : 141) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
POT15_HUMAN
|
||||||
| NC score | 0.019652 (rank : 142) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
FA21C_HUMAN
|
||||||
| NC score | 0.019636 (rank : 143) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
CT175_HUMAN
|
||||||
| NC score | 0.019325 (rank : 144) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96MK2, Q5QPB6, Q9NQQ2 | Gene names | C20orf175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf175. | |||||
|
Z3H7B_HUMAN
|
||||||
| NC score | 0.019172 (rank : 145) | θ value | 1.38821 (rank : 79) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGR2, Q5TFX9, Q8TBT9, Q9H8B6, Q9UGQ9, Q9UGR0, Q9UGR1, Q9UK03, Q9UPW9 | Gene names | ZC3H7B, KIAA1031 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7B (Rotavirus 'X'- associated non-structural protein) (RoXaN). | |||||
|
BRE1A_MOUSE
|
||||||
| NC score | 0.019143 (rank : 146) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
BRD7_HUMAN
|
||||||
| NC score | 0.018993 (rank : 147) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NPI1, Q4VC09, Q8N2L9, Q96KA4, Q9BV48, Q9UH59 | Gene names | BRD7, BP75, CELTIX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein) (Protein CELTIX-1). | |||||
|
F125A_MOUSE
|
||||||
| NC score | 0.018379 (rank : 148) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q78HU3 | Gene names | Fam125a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM125A. | |||||
|
HNF1B_HUMAN
|
||||||
| NC score | 0.018072 (rank : 149) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35680 | Gene names | TCF2, HNF1B | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-beta (HNF-1beta) (HNF-1B) (Variant hepatic nuclear factor 1) (VHNF1) (Homeoprotein LFB3) (Transcription factor 2) (TCF-2). | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.018022 (rank : 150) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
BRE1A_HUMAN
|
||||||
| NC score | 0.017784 (rank : 151) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
FGD5_HUMAN
|
||||||
| NC score | 0.017343 (rank : 152) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
K0423_HUMAN
|
||||||
| NC score | 0.017341 (rank : 153) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
DJBP_MOUSE
|
||||||
| NC score | 0.017308 (rank : 154) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P1E8, Q5DTV8, Q6PGK8, Q9D2D8 | Gene names | Djbp, Kiaa1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein. | |||||
|
SH2D3_HUMAN
|
||||||
| NC score | 0.016557 (rank : 155) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N5H7, Q5HYE5, Q6UY42, Q8N6X3, Q9Y2X5 | Gene names | SH2D3C, NSP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (Novel SH2-containing protein 3). | |||||
|
CJ006_MOUSE
|
||||||
| NC score | 0.015698 (rank : 156) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P9P0, Q8C041, Q8C0J5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf6 homolog. | |||||
|
CDKL5_HUMAN
|
||||||
| NC score | 0.015357 (rank : 157) | θ value | 0.00869519 (rank : 12) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.014949 (rank : 158) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
DACT1_HUMAN
|
||||||
| NC score | 0.014451 (rank : 159) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYF0, Q86TY0 | Gene names | DACT1, DPR1, HNG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (hDPR1) (Heptacellular carcinoma novel gene 3 protein). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.013884 (rank : 160) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.013843 (rank : 161) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.013752 (rank : 162) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.013696 (rank : 163) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.013635 (rank : 164) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.013333 (rank : 165) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.013330 (rank : 166) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
EGR1_MOUSE
|
||||||
| NC score | 0.012675 (rank : 167) | θ value | 0.0330416 (rank : 15) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
MCCA_HUMAN
|
||||||
| NC score | 0.012165 (rank : 168) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96RQ3, Q59ES4, Q9H959, Q9NS97 | Gene names | MCCC1, MCCA | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
CO4A2_HUMAN
|
||||||
| NC score | 0.011938 (rank : 169) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
DIAP1_MOUSE
|
||||||
| NC score | 0.011724 (rank : 170) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | 0.011606 (rank : 171) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
ENAH_HUMAN
|
||||||
| NC score | 0.011389 (rank : 172) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
DMXL1_MOUSE
|
||||||
| NC score | 0.010890 (rank : 173) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PNC0, Q3TLG8, Q6PHM6, Q8BQF2 | Gene names | Dmxl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
CD248_MOUSE
|
||||||
| NC score | 0.009997 (rank : 174) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
DDX5_HUMAN
|
||||||
| NC score | 0.009843 (rank : 175) | θ value | 0.813845 (rank : 62) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
EGR1_HUMAN
|
||||||
| NC score | 0.008846 (rank : 176) | θ value | 0.279714 (rank : 39) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18146 | Gene names | EGR1, ZNF225 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A) (Transcription factor ETR103) (Zinc finger protein 225) (AT225). | |||||
|
K1C9_HUMAN
|
||||||
| NC score | 0.008362 (rank : 177) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35527, O00109, Q14665 | Gene names | KRT9 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 9 (Cytokeratin-9) (CK-9) (Keratin-9) (K9). | |||||
|
DDX5_MOUSE
|
||||||
| NC score | 0.008270 (rank : 178) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
B3A3_MOUSE
|
||||||
| NC score | 0.008218 (rank : 179) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16283 | Gene names | Slc4a3, Ae3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3). | |||||
|
T240L_HUMAN
|
||||||
| NC score | 0.007623 (rank : 180) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q71F56, Q68DN4, Q9H8C0, Q9NSY9, Q9UFD8, Q9UPX5 | Gene names | THRAP2, KIAA1025, PROSIT240, TRAP240L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.007538 (rank : 181) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.005761 (rank : 182) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
LPHN1_HUMAN
|
||||||
| NC score | 0.005705 (rank : 183) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94910, Q96IE7, Q9BU07, Q9HAR3 | Gene names | LPHN1, KIAA0821, LEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-1 precursor (Calcium-independent alpha-latrotoxin receptor 1) (Lectomedin-2). | |||||
|
SMAD1_MOUSE
|
||||||
| NC score | 0.005601 (rank : 184) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70340, P70442, Q9CYK6 | Gene names | Smad1, Madh1, Madr1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Mothers-against-DPP-related 1) (mMad1) (Dwarfin-A) (Dwf-A). | |||||
|
SOX8_MOUSE
|
||||||
| NC score | 0.005529 (rank : 185) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q04886 | Gene names | Sox8, Sox-8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
MEFV_HUMAN
|
||||||
| NC score | 0.005385 (rank : 186) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.005235 (rank : 187) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CAC1F_HUMAN
|
||||||
| NC score | 0.004189 (rank : 188) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
LATS2_MOUSE
|
||||||
| NC score | 0.003854 (rank : 189) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TSJ6, Q8CDJ4, Q9JMI3 | Gene names | Lats2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS2 (EC 2.7.11.1) (Large tumor suppressor homolog 2) (Serine/threonine-protein kinase kpm) (Kinase phosphorylated during mitosis protein). | |||||
|
STK6_HUMAN
|
||||||
| NC score | 0.003277 (rank : 190) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14965, O60445, O75873, Q9BQD6, Q9UPG5 | Gene names | STK6, AIK, ARK1, AURA, BTAK, STK15 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 6 (EC 2.7.11.1) (Serine/threonine kinase 15) (Aurora/IPL1-related kinase 1) (Aurora-related kinase 1) (hARK1) (Aurora-A) (Breast-tumor-amplified kinase). | |||||
|
CDK8_HUMAN
|
||||||
| NC score | 0.001865 (rank : 191) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49336 | Gene names | CDK8 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division protein kinase 8 (EC 2.7.11.22) (EC 2.7.11.23) (Protein kinase K35). | |||||
|
PTN14_MOUSE
|
||||||
| NC score | 0.001265 (rank : 192) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62130 | Gene names | Ptpn14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP36). | |||||
|
PRDM1_MOUSE
|
||||||
| NC score | 0.000830 (rank : 193) | θ value | 1.38821 (rank : 78) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 766 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60636 | Gene names | Prdm1, Blimp1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 1 (PR domain-containing protein 1) (Beta-interferon gene positive-regulatory domain I-binding factor) (BLIMP-1). | |||||
|
NOTC3_MOUSE
|
||||||
| NC score | 0.000170 (rank : 194) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
PCD17_HUMAN
|
||||||
| NC score | -0.000258 (rank : 195) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 168 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14917 | Gene names | PCDH17, PCDH68, PCH68 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-17 precursor (Protocadherin-68). | |||||