Please be patient as the page loads
|
GDIB_MOUSE
|
||||||
| SwissProt Accessions | Q61598, Q541Z9, Q8C530, Q9D8M9 | Gene names | Gdi2, Gdi3 | |||
|
Domain Architecture |
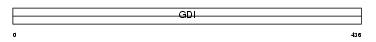
|
|||||
| Description | Rab GDP dissociation inhibitor beta (Rab GDI beta) (Guanosine diphosphate dissociation inhibitor 2) (GDI-2) (GDI-3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GDIA_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998135 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P31150, P50394, Q7Z2G6, Q7Z2G9, Q7Z2H5, Q7Z2I6 | Gene names | GDI1, OPHN2, RABGDIA, XAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rab GDP dissociation inhibitor alpha (Rab GDI alpha) (Guanosine diphosphate dissociation inhibitor 1) (GDI-1) (XAP-4) (Oligophrenin- 2). | |||||
|
GDIA_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998074 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P50396, Q8VHM3, Q91Y71, Q96CX5 | Gene names | Gdi1, Rabgdia | |||
|
Domain Architecture |
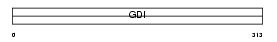
|
|||||
| Description | Rab GDP dissociation inhibitor alpha (Rab GDI alpha) (Guanosine diphosphate dissociation inhibitor 1) (GDI-1). | |||||
|
GDIB_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999140 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P50395, O43928, Q9UQM6 | Gene names | GDI2, RABGDIB | |||
|
Domain Architecture |
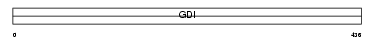
|
|||||
| Description | Rab GDP dissociation inhibitor beta (Rab GDI beta) (Guanosine diphosphate dissociation inhibitor 2) (GDI-2). | |||||
|
GDIB_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61598, Q541Z9, Q8C530, Q9D8M9 | Gene names | Gdi2, Gdi3 | |||
|
Domain Architecture |

|
|||||
| Description | Rab GDP dissociation inhibitor beta (Rab GDI beta) (Guanosine diphosphate dissociation inhibitor 2) (GDI-2) (GDI-3). | |||||
|
RAE2_HUMAN
|
||||||
| θ value | 1.2049e-28 (rank : 5) | NC score | 0.712696 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P26374, Q9H1Y4 | Gene names | CHML, REP2 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 2 (Rab escort protein 2) (REP-2) (Choroideraemia-like protein). | |||||
|
RAE2_MOUSE
|
||||||
| θ value | 5.59698e-26 (rank : 6) | NC score | 0.697923 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QZD5, Q8CH16 | Gene names | Chml, Rep2 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 2 (Rab escort protein 2) (REP-2) (Choroideraemia-like protein). | |||||
|
RAE1_HUMAN
|
||||||
| θ value | 8.08199e-25 (rank : 7) | NC score | 0.693832 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P24386, O43732 | Gene names | CHM, REP1, TCD | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 1 (Rab escort protein 1) (REP-1) (Choroideraemia protein) (TCD protein). | |||||
|
RAE1_MOUSE
|
||||||
| θ value | 1.05554e-24 (rank : 8) | NC score | 0.693838 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QXG2, Q80UV8 | Gene names | Chm, Rep1 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 1 (Rab escort protein 1) (REP-1) (Choroideraemia protein homolog). | |||||
|
LRCH2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 9) | NC score | 0.027550 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UMG5, Q7TNP9, Q80TC9 | Gene names | Lrch2, Kiaa1495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 2. | |||||
|
LRCH2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 10) | NC score | 0.023673 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VUJ6, Q9HA88, Q9P233 | Gene names | LRCH2, KIAA1495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 2. | |||||
|
MYCN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.060205 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
PEX6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 12) | NC score | 0.011452 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
RETST_HUMAN
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.083943 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6NUM9, Q53R95, Q53SA9, Q6UX05, Q8N2H5, Q96FA4, Q9NXE5 | Gene names | RETSAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
MYCN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.037273 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
SAP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 15) | NC score | 0.024631 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61207, Q60861, Q64006, Q64219 | Gene names | Psap, Sgp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sulfated glycoprotein 1 precursor (SGP-1) (Prosaposin). | |||||
|
BAG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 16) | NC score | 0.011893 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99933, O75315, Q14414, Q9Y2V4 | Gene names | BAG1 | |||
|
Domain Architecture |
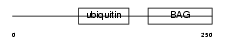
|
|||||
| Description | BAG family molecular chaperone regulator 1 (BCL-2-binding athanogene- 1) (BAG-1) (Glucocorticoid receptor-associated protein RAP46). | |||||
|
RETST_MOUSE
|
||||||
| θ value | 8.99809 (rank : 17) | NC score | 0.054691 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64FW2, Q3UNN9, Q78JX8, Q8VHE7, Q9CW85 | Gene names | Retsat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
GDIB_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61598, Q541Z9, Q8C530, Q9D8M9 | Gene names | Gdi2, Gdi3 | |||
|
Domain Architecture |

|
|||||
| Description | Rab GDP dissociation inhibitor beta (Rab GDI beta) (Guanosine diphosphate dissociation inhibitor 2) (GDI-2) (GDI-3). | |||||
|
GDIB_HUMAN
|
||||||
| NC score | 0.999140 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P50395, O43928, Q9UQM6 | Gene names | GDI2, RABGDIB | |||
|
Domain Architecture |

|
|||||
| Description | Rab GDP dissociation inhibitor beta (Rab GDI beta) (Guanosine diphosphate dissociation inhibitor 2) (GDI-2). | |||||
|
GDIA_HUMAN
|
||||||
| NC score | 0.998135 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P31150, P50394, Q7Z2G6, Q7Z2G9, Q7Z2H5, Q7Z2I6 | Gene names | GDI1, OPHN2, RABGDIA, XAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rab GDP dissociation inhibitor alpha (Rab GDI alpha) (Guanosine diphosphate dissociation inhibitor 1) (GDI-1) (XAP-4) (Oligophrenin- 2). | |||||
|
GDIA_MOUSE
|
||||||
| NC score | 0.998074 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P50396, Q8VHM3, Q91Y71, Q96CX5 | Gene names | Gdi1, Rabgdia | |||
|
Domain Architecture |

|
|||||
| Description | Rab GDP dissociation inhibitor alpha (Rab GDI alpha) (Guanosine diphosphate dissociation inhibitor 1) (GDI-1). | |||||
|
RAE2_HUMAN
|
||||||
| NC score | 0.712696 (rank : 5) | θ value | 1.2049e-28 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P26374, Q9H1Y4 | Gene names | CHML, REP2 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 2 (Rab escort protein 2) (REP-2) (Choroideraemia-like protein). | |||||
|
RAE2_MOUSE
|
||||||
| NC score | 0.697923 (rank : 6) | θ value | 5.59698e-26 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QZD5, Q8CH16 | Gene names | Chml, Rep2 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 2 (Rab escort protein 2) (REP-2) (Choroideraemia-like protein). | |||||
|
RAE1_MOUSE
|
||||||
| NC score | 0.693838 (rank : 7) | θ value | 1.05554e-24 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QXG2, Q80UV8 | Gene names | Chm, Rep1 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 1 (Rab escort protein 1) (REP-1) (Choroideraemia protein homolog). | |||||
|
RAE1_HUMAN
|
||||||
| NC score | 0.693832 (rank : 8) | θ value | 8.08199e-25 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P24386, O43732 | Gene names | CHM, REP1, TCD | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 1 (Rab escort protein 1) (REP-1) (Choroideraemia protein) (TCD protein). | |||||
|
RETST_HUMAN
|
||||||
| NC score | 0.083943 (rank : 9) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6NUM9, Q53R95, Q53SA9, Q6UX05, Q8N2H5, Q96FA4, Q9NXE5 | Gene names | RETSAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
MYCN_HUMAN
|
||||||
| NC score | 0.060205 (rank : 10) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
RETST_MOUSE
|
||||||
| NC score | 0.054691 (rank : 11) | θ value | 8.99809 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64FW2, Q3UNN9, Q78JX8, Q8VHE7, Q9CW85 | Gene names | Retsat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
MYCN_MOUSE
|
||||||
| NC score | 0.037273 (rank : 12) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
LRCH2_MOUSE
|
||||||
| NC score | 0.027550 (rank : 13) | θ value | 0.279714 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UMG5, Q7TNP9, Q80TC9 | Gene names | Lrch2, Kiaa1495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 2. | |||||
|
SAP_MOUSE
|
||||||
| NC score | 0.024631 (rank : 14) | θ value | 5.27518 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61207, Q60861, Q64006, Q64219 | Gene names | Psap, Sgp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sulfated glycoprotein 1 precursor (SGP-1) (Prosaposin). | |||||
|
LRCH2_HUMAN
|
||||||
| NC score | 0.023673 (rank : 15) | θ value | 0.365318 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VUJ6, Q9HA88, Q9P233 | Gene names | LRCH2, KIAA1495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 2. | |||||
|
BAG1_HUMAN
|
||||||
| NC score | 0.011893 (rank : 16) | θ value | 8.99809 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99933, O75315, Q14414, Q9Y2V4 | Gene names | BAG1 | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 1 (BCL-2-binding athanogene- 1) (BAG-1) (Glucocorticoid receptor-associated protein RAP46). | |||||
|
PEX6_HUMAN
|
||||||
| NC score | 0.011452 (rank : 17) | θ value | 3.0926 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||