Please be patient as the page loads
|
RETST_HUMAN
|
||||||
| SwissProt Accessions | Q6NUM9, Q53R95, Q53SA9, Q6UX05, Q8N2H5, Q96FA4, Q9NXE5 | Gene names | RETSAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RETST_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6NUM9, Q53R95, Q53SA9, Q6UX05, Q8N2H5, Q96FA4, Q9NXE5 | Gene names | RETSAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
RETST_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995511 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q64FW2, Q3UNN9, Q78JX8, Q8VHE7, Q9CW85 | Gene names | Retsat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
CJ033_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 3) | NC score | 0.392101 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3U4I7, Q8CAN2, Q9D630 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 homolog (EC 1.-.-.-). | |||||
|
CJ033_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 4) | NC score | 0.342342 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N2H3, Q5TAA9, Q9BRQ1 | Gene names | C10orf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 (EC 1.-.-.-). | |||||
|
AOFB_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 5) | NC score | 0.191881 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27338, Q7Z6S2 | Gene names | MAOB | |||
|
Domain Architecture |
|
|||||
| Description | Amine oxidase [flavin-containing] B (EC 1.4.3.4) (Monoamine oxidase type B) (MAO-B). | |||||
|
AOFA_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 6) | NC score | 0.182123 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64133, Q8K0Z8 | Gene names | Maoa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A). | |||||
|
RAE1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 7) | NC score | 0.164207 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P24386, O43732 | Gene names | CHM, REP1, TCD | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 1 (Rab escort protein 1) (REP-1) (Choroideraemia protein) (TCD protein). | |||||
|
RAE2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 8) | NC score | 0.163314 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P26374, Q9H1Y4 | Gene names | CHML, REP2 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 2 (Rab escort protein 2) (REP-2) (Choroideraemia-like protein). | |||||
|
RAE2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 9) | NC score | 0.164941 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QZD5, Q8CH16 | Gene names | Chml, Rep2 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 2 (Rab escort protein 2) (REP-2) (Choroideraemia-like protein). | |||||
|
RAE1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 10) | NC score | 0.159380 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QXG2, Q80UV8 | Gene names | Chm, Rep1 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 1 (Rab escort protein 1) (REP-1) (Choroideraemia protein homolog). | |||||
|
DLDH_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.120903 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08749 | Gene names | Dld | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase). | |||||
|
DLDH_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 12) | NC score | 0.118015 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P09622, Q14131, Q14167 | Gene names | DLD, GCSL, LAD, PHE3 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase) (Glycine cleavage system L protein). | |||||
|
ERG1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.101234 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P52019 | Gene names | Sqle, Erg1 | |||
|
Domain Architecture |
|
|||||
| Description | Squalene monooxygenase (EC 1.14.99.7) (Squalene epoxidase) (SE). | |||||
|
PPOX_HUMAN
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.090024 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P50336 | Gene names | PPOX | |||
|
Domain Architecture |
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
AOFB_MOUSE
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.126394 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BW75, Q8C0B2 | Gene names | Maob | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amine oxidase [flavin-containing] B (EC 1.4.3.4) (Monoamine oxidase type B) (MAO-B). | |||||
|
TRXR2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.060782 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JLT4, Q91YX4, Q9JHA7, Q9JMH5 | Gene names | Txnrd2, Trxr2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 2, mitochondrial precursor (EC 1.8.1.9) (TR3). | |||||
|
SOX_HUMAN
|
||||||
| θ value | 1.81305 (rank : 17) | NC score | 0.086542 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P0Z9, Q96H28, Q9C070 | Gene names | PIPOX, LPIPOX, PSO | |||
|
Domain Architecture |
|
|||||
| Description | Peroxisomal sarcosine oxidase (EC 1.5.3.1) (PSO) (L-pipecolate oxidase) (EC 1.5.3.7) (L-pipecolic acid oxidase). | |||||
|
ERG1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.076680 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14534, Q9UEK6 | Gene names | SQLE, ERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Squalene monooxygenase (EC 1.14.99.7) (Squalene epoxidase) (SE). | |||||
|
GDIB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.083943 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61598, Q541Z9, Q8C530, Q9D8M9 | Gene names | Gdi2, Gdi3 | |||
|
Domain Architecture |
|
|||||
| Description | Rab GDP dissociation inhibitor beta (Rab GDI beta) (Guanosine diphosphate dissociation inhibitor 2) (GDI-2) (GDI-3). | |||||
|
M2GD_MOUSE
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.043410 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DBT9, Q8R1S7 | Gene names | Dmgdh | |||
|
Domain Architecture |

|
|||||
| Description | Dimethylglycine dehydrogenase, mitochondrial precursor (EC 1.5.99.2) (ME2GLYDH). | |||||
|
SOX_MOUSE
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.075442 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D826, O55223 | Gene names | Pipox, Pso | |||
|
Domain Architecture |
|
|||||
| Description | Peroxisomal sarcosine oxidase (EC 1.5.3.1) (PSO) (L-pipecolate oxidase) (EC 1.5.3.7) (L-pipecolic acid oxidase). | |||||
|
ETFD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.030674 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q921G7, Q3U7K2, Q8BK82, Q9DCT9 | Gene names | Etfdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial precursor (EC 1.5.5.1) (ETF-QO) (ETF-ubiquinone oxidoreductase) (ETF dehydrogenase) (Electron-transferring- flavoprotein dehydrogenase). | |||||
|
M2GD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.034827 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UI17 | Gene names | DMGDH | |||
|
Domain Architecture |

|
|||||
| Description | Dimethylglycine dehydrogenase, mitochondrial precursor (EC 1.5.99.2) (ME2GLYDH). | |||||
|
GDIA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.073378 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P31150, P50394, Q7Z2G6, Q7Z2G9, Q7Z2H5, Q7Z2I6 | Gene names | GDI1, OPHN2, RABGDIA, XAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rab GDP dissociation inhibitor alpha (Rab GDI alpha) (Guanosine diphosphate dissociation inhibitor 1) (GDI-1) (XAP-4) (Oligophrenin- 2). | |||||
|
GDIA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.073598 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P50396, Q8VHM3, Q91Y71, Q96CX5 | Gene names | Gdi1, Rabgdia | |||
|
Domain Architecture |
|
|||||
| Description | Rab GDP dissociation inhibitor alpha (Rab GDI alpha) (Guanosine diphosphate dissociation inhibitor 1) (GDI-1). | |||||
|
GDIB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.073145 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P50395, O43928, Q9UQM6 | Gene names | GDI2, RABGDIB | |||
|
Domain Architecture |
|
|||||
| Description | Rab GDP dissociation inhibitor beta (Rab GDI beta) (Guanosine diphosphate dissociation inhibitor 2) (GDI-2). | |||||
|
LTBP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.002731 (rank : 28) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
AOFA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.107054 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21397, Q16426 | Gene names | MAOA | |||
|
Domain Architecture |

|
|||||
| Description | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A). | |||||
|
RETST_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6NUM9, Q53R95, Q53SA9, Q6UX05, Q8N2H5, Q96FA4, Q9NXE5 | Gene names | RETSAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
RETST_MOUSE
|
||||||
| NC score | 0.995511 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q64FW2, Q3UNN9, Q78JX8, Q8VHE7, Q9CW85 | Gene names | Retsat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
CJ033_MOUSE
|
||||||
| NC score | 0.392101 (rank : 3) | θ value | 6.87365e-08 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3U4I7, Q8CAN2, Q9D630 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 homolog (EC 1.-.-.-). | |||||
|
CJ033_HUMAN
|
||||||
| NC score | 0.342342 (rank : 4) | θ value | 8.40245e-06 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N2H3, Q5TAA9, Q9BRQ1 | Gene names | C10orf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 (EC 1.-.-.-). | |||||
|
AOFB_HUMAN
|
||||||
| NC score | 0.191881 (rank : 5) | θ value | 9.29e-05 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27338, Q7Z6S2 | Gene names | MAOB | |||
|
Domain Architecture |
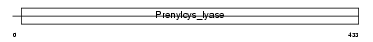
|
|||||
| Description | Amine oxidase [flavin-containing] B (EC 1.4.3.4) (Monoamine oxidase type B) (MAO-B). | |||||
|
AOFA_MOUSE
|
||||||
| NC score | 0.182123 (rank : 6) | θ value | 0.000786445 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64133, Q8K0Z8 | Gene names | Maoa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A). | |||||
|
RAE2_MOUSE
|
||||||
| NC score | 0.164941 (rank : 7) | θ value | 0.0252991 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QZD5, Q8CH16 | Gene names | Chml, Rep2 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 2 (Rab escort protein 2) (REP-2) (Choroideraemia-like protein). | |||||
|
RAE1_HUMAN
|
||||||
| NC score | 0.164207 (rank : 8) | θ value | 0.0252991 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P24386, O43732 | Gene names | CHM, REP1, TCD | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 1 (Rab escort protein 1) (REP-1) (Choroideraemia protein) (TCD protein). | |||||
|
RAE2_HUMAN
|
||||||
| NC score | 0.163314 (rank : 9) | θ value | 0.0252991 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P26374, Q9H1Y4 | Gene names | CHML, REP2 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 2 (Rab escort protein 2) (REP-2) (Choroideraemia-like protein). | |||||
|
RAE1_MOUSE
|
||||||
| NC score | 0.159380 (rank : 10) | θ value | 0.0431538 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QXG2, Q80UV8 | Gene names | Chm, Rep1 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 1 (Rab escort protein 1) (REP-1) (Choroideraemia protein homolog). | |||||
|
AOFB_MOUSE
|
||||||
| NC score | 0.126394 (rank : 11) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BW75, Q8C0B2 | Gene names | Maob | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amine oxidase [flavin-containing] B (EC 1.4.3.4) (Monoamine oxidase type B) (MAO-B). | |||||
|
DLDH_MOUSE
|
||||||
| NC score | 0.120903 (rank : 12) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08749 | Gene names | Dld | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase). | |||||
|
DLDH_HUMAN
|
||||||
| NC score | 0.118015 (rank : 13) | θ value | 0.0736092 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P09622, Q14131, Q14167 | Gene names | DLD, GCSL, LAD, PHE3 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase) (Glycine cleavage system L protein). | |||||
|
AOFA_HUMAN
|
||||||
| NC score | 0.107054 (rank : 14) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21397, Q16426 | Gene names | MAOA | |||
|
Domain Architecture |

|
|||||
| Description | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A). | |||||
|
ERG1_MOUSE
|
||||||
| NC score | 0.101234 (rank : 15) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P52019 | Gene names | Sqle, Erg1 | |||
|
Domain Architecture |

|
|||||
| Description | Squalene monooxygenase (EC 1.14.99.7) (Squalene epoxidase) (SE). | |||||
|
PPOX_HUMAN
|
||||||
| NC score | 0.090024 (rank : 16) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P50336 | Gene names | PPOX | |||
|
Domain Architecture |
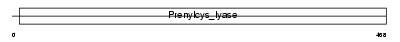
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
SOX_HUMAN
|
||||||
| NC score | 0.086542 (rank : 17) | θ value | 1.81305 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P0Z9, Q96H28, Q9C070 | Gene names | PIPOX, LPIPOX, PSO | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal sarcosine oxidase (EC 1.5.3.1) (PSO) (L-pipecolate oxidase) (EC 1.5.3.7) (L-pipecolic acid oxidase). | |||||
|
GDIB_MOUSE
|
||||||
| NC score | 0.083943 (rank : 18) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61598, Q541Z9, Q8C530, Q9D8M9 | Gene names | Gdi2, Gdi3 | |||
|
Domain Architecture |

|
|||||
| Description | Rab GDP dissociation inhibitor beta (Rab GDI beta) (Guanosine diphosphate dissociation inhibitor 2) (GDI-2) (GDI-3). | |||||
|
ERG1_HUMAN
|
||||||
| NC score | 0.076680 (rank : 19) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14534, Q9UEK6 | Gene names | SQLE, ERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Squalene monooxygenase (EC 1.14.99.7) (Squalene epoxidase) (SE). | |||||
|
SOX_MOUSE
|
||||||
| NC score | 0.075442 (rank : 20) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D826, O55223 | Gene names | Pipox, Pso | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal sarcosine oxidase (EC 1.5.3.1) (PSO) (L-pipecolate oxidase) (EC 1.5.3.7) (L-pipecolic acid oxidase). | |||||
|
GDIA_MOUSE
|
||||||
| NC score | 0.073598 (rank : 21) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P50396, Q8VHM3, Q91Y71, Q96CX5 | Gene names | Gdi1, Rabgdia | |||
|
Domain Architecture |

|
|||||
| Description | Rab GDP dissociation inhibitor alpha (Rab GDI alpha) (Guanosine diphosphate dissociation inhibitor 1) (GDI-1). | |||||
|
GDIA_HUMAN
|
||||||
| NC score | 0.073378 (rank : 22) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P31150, P50394, Q7Z2G6, Q7Z2G9, Q7Z2H5, Q7Z2I6 | Gene names | GDI1, OPHN2, RABGDIA, XAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rab GDP dissociation inhibitor alpha (Rab GDI alpha) (Guanosine diphosphate dissociation inhibitor 1) (GDI-1) (XAP-4) (Oligophrenin- 2). | |||||
|
GDIB_HUMAN
|
||||||
| NC score | 0.073145 (rank : 23) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P50395, O43928, Q9UQM6 | Gene names | GDI2, RABGDIB | |||
|
Domain Architecture |

|
|||||
| Description | Rab GDP dissociation inhibitor beta (Rab GDI beta) (Guanosine diphosphate dissociation inhibitor 2) (GDI-2). | |||||
|
TRXR2_MOUSE
|
||||||
| NC score | 0.060782 (rank : 24) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JLT4, Q91YX4, Q9JHA7, Q9JMH5 | Gene names | Txnrd2, Trxr2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 2, mitochondrial precursor (EC 1.8.1.9) (TR3). | |||||
|
M2GD_MOUSE
|
||||||
| NC score | 0.043410 (rank : 25) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DBT9, Q8R1S7 | Gene names | Dmgdh | |||
|
Domain Architecture |

|
|||||
| Description | Dimethylglycine dehydrogenase, mitochondrial precursor (EC 1.5.99.2) (ME2GLYDH). | |||||
|
M2GD_HUMAN
|
||||||
| NC score | 0.034827 (rank : 26) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UI17 | Gene names | DMGDH | |||
|
Domain Architecture |

|
|||||
| Description | Dimethylglycine dehydrogenase, mitochondrial precursor (EC 1.5.99.2) (ME2GLYDH). | |||||
|
ETFD_MOUSE
|
||||||
| NC score | 0.030674 (rank : 27) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q921G7, Q3U7K2, Q8BK82, Q9DCT9 | Gene names | Etfdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial precursor (EC 1.5.5.1) (ETF-QO) (ETF-ubiquinone oxidoreductase) (ETF dehydrogenase) (Electron-transferring- flavoprotein dehydrogenase). | |||||
|
LTBP2_HUMAN
|
||||||
| NC score | 0.002731 (rank : 28) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||