Please be patient as the page loads
|
ERG1_HUMAN
|
||||||
| SwissProt Accessions | Q14534, Q9UEK6 | Gene names | SQLE, ERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Squalene monooxygenase (EC 1.14.99.7) (Squalene epoxidase) (SE). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ERG1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14534, Q9UEK6 | Gene names | SQLE, ERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Squalene monooxygenase (EC 1.14.99.7) (Squalene epoxidase) (SE). | |||||
|
ERG1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994500 (rank : 2) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P52019 | Gene names | Sqle, Erg1 | |||
|
Domain Architecture |
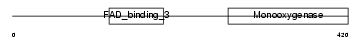
|
|||||
| Description | Squalene monooxygenase (EC 1.14.99.7) (Squalene epoxidase) (SE). | |||||
|
OXLA_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 3) | NC score | 0.149275 (rank : 3) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96RQ9, Q4GZN1, Q4GZN2, Q6P2Q3, Q8TEM5, Q96RQ8 | Gene names | IL4I1, FIG1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (hFIG1). | |||||
|
KMO_HUMAN
|
||||||
| θ value | 0.163984 (rank : 4) | NC score | 0.100234 (rank : 5) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15229, Q5SY07, Q5SY08, Q5SY09 | Gene names | KMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kynurenine 3-monooxygenase (EC 1.14.13.9) (Kynurenine 3-hydroxylase). | |||||
|
OXLA_MOUSE
|
||||||
| θ value | 0.279714 (rank : 5) | NC score | 0.135881 (rank : 4) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O09046, Q3T9N9, Q3U7S6, Q3V0K2, Q6Y632, Q6YBV6, Q6YDI8, Q8R2G8, Q9CXK7 | Gene names | Il4i1, Fig1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (mFIG1). | |||||
|
AOF1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 6) | NC score | 0.069695 (rank : 9) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NB78, Q5TGV3, Q6AI15, Q6ZUU4, Q8N258, Q96EL7 | Gene names | AOF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
AOF1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 7) | NC score | 0.069934 (rank : 8) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CIG3, Q8C5C4, Q8CEC1 | Gene names | Aof1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
CM007_MOUSE
|
||||||
| θ value | 1.06291 (rank : 8) | NC score | 0.043105 (rank : 12) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
DLDH_HUMAN
|
||||||
| θ value | 2.36792 (rank : 9) | NC score | 0.054939 (rank : 11) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P09622, Q14131, Q14167 | Gene names | DLD, GCSL, LAD, PHE3 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase) (Glycine cleavage system L protein). | |||||
|
DLDH_MOUSE
|
||||||
| θ value | 2.36792 (rank : 10) | NC score | 0.054989 (rank : 10) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08749 | Gene names | Dld | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase). | |||||
|
RETST_HUMAN
|
||||||
| θ value | 2.36792 (rank : 11) | NC score | 0.076680 (rank : 6) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6NUM9, Q53R95, Q53SA9, Q6UX05, Q8N2H5, Q96FA4, Q9NXE5 | Gene names | RETSAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
RETST_MOUSE
|
||||||
| θ value | 2.36792 (rank : 12) | NC score | 0.075771 (rank : 7) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64FW2, Q3UNN9, Q78JX8, Q8VHE7, Q9CW85 | Gene names | Retsat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
CM007_HUMAN
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.037001 (rank : 13) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5W0B1, Q8TBY2, Q9H0T2, Q9H8M0 | Gene names | C13orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7. | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 14) | NC score | 0.009049 (rank : 18) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
HEAT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 15) | NC score | 0.025271 (rank : 16) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86Y56, Q69YL1, Q96FI9, Q9NX75 | Gene names | HEATR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HEAT repeat-containing protein 2. | |||||
|
K0586_HUMAN
|
||||||
| θ value | 6.88961 (rank : 16) | NC score | 0.018760 (rank : 17) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BVV6, O60328 | Gene names | KIAA0586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0586. | |||||
|
COQ6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 17) | NC score | 0.026049 (rank : 15) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2Z9, Q96CA1, Q96CK2 | Gene names | COQ6 | |||
|
Domain Architecture |
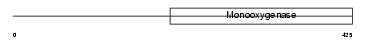
|
|||||
| Description | Ubiquinone biosynthesis monooxygenase COQ6 (EC 1.14.13.-). | |||||
|
COQ6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 18) | NC score | 0.026155 (rank : 14) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R1S0 | Gene names | Coq6 | |||
|
Domain Architecture |
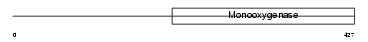
|
|||||
| Description | Ubiquinone biosynthesis monooxygenase COQ6 (EC 1.14.13.-). | |||||
|
PLK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 19) | NC score | 0.000538 (rank : 19) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07832 | Gene names | Plk1, Plk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK1 (EC 2.7.11.21) (Polo-like kinase 1) (PLK-1) (Serine/threonine-protein kinase 13) (STPK13). | |||||
|
ERG1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14534, Q9UEK6 | Gene names | SQLE, ERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Squalene monooxygenase (EC 1.14.99.7) (Squalene epoxidase) (SE). | |||||
|
ERG1_MOUSE
|
||||||
| NC score | 0.994500 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P52019 | Gene names | Sqle, Erg1 | |||
|
Domain Architecture |

|
|||||
| Description | Squalene monooxygenase (EC 1.14.99.7) (Squalene epoxidase) (SE). | |||||
|
OXLA_HUMAN
|
||||||
| NC score | 0.149275 (rank : 3) | θ value | 0.0961366 (rank : 3) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96RQ9, Q4GZN1, Q4GZN2, Q6P2Q3, Q8TEM5, Q96RQ8 | Gene names | IL4I1, FIG1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (hFIG1). | |||||
|
OXLA_MOUSE
|
||||||
| NC score | 0.135881 (rank : 4) | θ value | 0.279714 (rank : 5) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O09046, Q3T9N9, Q3U7S6, Q3V0K2, Q6Y632, Q6YBV6, Q6YDI8, Q8R2G8, Q9CXK7 | Gene names | Il4i1, Fig1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (mFIG1). | |||||
|
KMO_HUMAN
|
||||||
| NC score | 0.100234 (rank : 5) | θ value | 0.163984 (rank : 4) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15229, Q5SY07, Q5SY08, Q5SY09 | Gene names | KMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kynurenine 3-monooxygenase (EC 1.14.13.9) (Kynurenine 3-hydroxylase). | |||||
|
RETST_HUMAN
|
||||||
| NC score | 0.076680 (rank : 6) | θ value | 2.36792 (rank : 11) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6NUM9, Q53R95, Q53SA9, Q6UX05, Q8N2H5, Q96FA4, Q9NXE5 | Gene names | RETSAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
RETST_MOUSE
|
||||||
| NC score | 0.075771 (rank : 7) | θ value | 2.36792 (rank : 12) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64FW2, Q3UNN9, Q78JX8, Q8VHE7, Q9CW85 | Gene names | Retsat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
AOF1_MOUSE
|
||||||
| NC score | 0.069934 (rank : 8) | θ value | 0.365318 (rank : 7) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CIG3, Q8C5C4, Q8CEC1 | Gene names | Aof1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
AOF1_HUMAN
|
||||||
| NC score | 0.069695 (rank : 9) | θ value | 0.365318 (rank : 6) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NB78, Q5TGV3, Q6AI15, Q6ZUU4, Q8N258, Q96EL7 | Gene names | AOF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
DLDH_MOUSE
|
||||||
| NC score | 0.054989 (rank : 10) | θ value | 2.36792 (rank : 10) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08749 | Gene names | Dld | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase). | |||||
|
DLDH_HUMAN
|
||||||
| NC score | 0.054939 (rank : 11) | θ value | 2.36792 (rank : 9) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P09622, Q14131, Q14167 | Gene names | DLD, GCSL, LAD, PHE3 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase) (Glycine cleavage system L protein). | |||||
|
CM007_MOUSE
|
||||||
| NC score | 0.043105 (rank : 12) | θ value | 1.06291 (rank : 8) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
CM007_HUMAN
|
||||||
| NC score | 0.037001 (rank : 13) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5W0B1, Q8TBY2, Q9H0T2, Q9H8M0 | Gene names | C13orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7. | |||||
|
COQ6_MOUSE
|
||||||
| NC score | 0.026155 (rank : 14) | θ value | 8.99809 (rank : 18) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R1S0 | Gene names | Coq6 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinone biosynthesis monooxygenase COQ6 (EC 1.14.13.-). | |||||
|
COQ6_HUMAN
|
||||||
| NC score | 0.026049 (rank : 15) | θ value | 8.99809 (rank : 17) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2Z9, Q96CA1, Q96CK2 | Gene names | COQ6 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquinone biosynthesis monooxygenase COQ6 (EC 1.14.13.-). | |||||
|
HEAT2_HUMAN
|
||||||
| NC score | 0.025271 (rank : 16) | θ value | 6.88961 (rank : 15) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86Y56, Q69YL1, Q96FI9, Q9NX75 | Gene names | HEATR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HEAT repeat-containing protein 2. | |||||
|
K0586_HUMAN
|
||||||
| NC score | 0.018760 (rank : 17) | θ value | 6.88961 (rank : 16) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BVV6, O60328 | Gene names | KIAA0586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0586. | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.009049 (rank : 18) | θ value | 5.27518 (rank : 14) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
PLK1_MOUSE
|
||||||
| NC score | 0.000538 (rank : 19) | θ value | 8.99809 (rank : 19) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07832 | Gene names | Plk1, Plk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK1 (EC 2.7.11.21) (Polo-like kinase 1) (PLK-1) (Serine/threonine-protein kinase 13) (STPK13). | |||||