Please be patient as the page loads
|
AOF1_HUMAN
|
||||||
| SwissProt Accessions | Q8NB78, Q5TGV3, Q6AI15, Q6ZUU4, Q8N258, Q96EL7 | Gene names | AOF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AOF1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8NB78, Q5TGV3, Q6AI15, Q6ZUU4, Q8N258, Q96EL7 | Gene names | AOF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
AOF1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.997324 (rank : 2) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CIG3, Q8C5C4, Q8CEC1 | Gene names | Aof1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
LSD1_HUMAN
|
||||||
| θ value | 1.46268e-50 (rank : 3) | NC score | 0.764986 (rank : 4) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
LSD1_MOUSE
|
||||||
| θ value | 1.46268e-50 (rank : 4) | NC score | 0.799463 (rank : 3) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6ZQ88, Q6PB53, Q8VEA1 | Gene names | Aof2, Kiaa0601, Lsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
PAOX_MOUSE
|
||||||
| θ value | 8.3442e-30 (rank : 5) | NC score | 0.759609 (rank : 5) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C0L6, Q8K254 | Gene names | Paox, Pao | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal N1-acetyl-spermine/spermidine oxidase (EC 1.5.3.11) (Polyamine oxidase). | |||||
|
SMOX_HUMAN
|
||||||
| θ value | 1.02001e-27 (rank : 6) | NC score | 0.729835 (rank : 6) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NWM0, Q5TE26, Q5TE27, Q6UY28, Q8IX00, Q96LC3, Q96LC4, Q96QT3, Q9BW38, Q9H6H1, Q9NP51, Q9NPY1, Q9NPY2 | Gene names | SMOX, C20orf16, SMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermine oxidase (EC 1.5.3.-) (Polyamine oxidase 1) (PAO-1) (PAOh1). | |||||
|
SMOX_MOUSE
|
||||||
| θ value | 1.92365e-26 (rank : 7) | NC score | 0.722427 (rank : 7) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99K82, Q70LA3, Q70LA4, Q70LA5, Q70LA7, Q70LA8, Q70LA9, Q70LB0, Q8CJ56, Q8CJ57 | Gene names | Smox, Smo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermine oxidase (EC 1.5.3.-) (Polyamine oxidase 1) (PAO-1) (PAOh1). | |||||
|
PAOX_HUMAN
|
||||||
| θ value | 3.07116e-24 (rank : 8) | NC score | 0.720134 (rank : 8) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6QHF9, Q6QHF5, Q6QHF6, Q6QHF7, Q6QHF8, Q6QHG0, Q6QHG1, Q6QHG2, Q6QHG3, Q6QHG4, Q6QHG5, Q6QHG6, Q86WP9, Q8N555, Q8NCX3 | Gene names | PAOX, PAO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal N1-acetyl-spermine/spermidine oxidase (EC 1.5.3.11) (Polyamine oxidase). | |||||
|
AOFB_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 9) | NC score | 0.575645 (rank : 9) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P27338, Q7Z6S2 | Gene names | MAOB | |||
|
Domain Architecture |
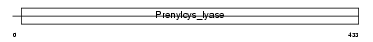
|
|||||
| Description | Amine oxidase [flavin-containing] B (EC 1.4.3.4) (Monoamine oxidase type B) (MAO-B). | |||||
|
AOFB_MOUSE
|
||||||
| θ value | 1.058e-16 (rank : 10) | NC score | 0.553924 (rank : 10) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BW75, Q8C0B2 | Gene names | Maob | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amine oxidase [flavin-containing] B (EC 1.4.3.4) (Monoamine oxidase type B) (MAO-B). | |||||
|
AOFA_MOUSE
|
||||||
| θ value | 1.52774e-15 (rank : 11) | NC score | 0.547626 (rank : 11) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q64133, Q8K0Z8 | Gene names | Maoa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A). | |||||
|
AOFA_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 12) | NC score | 0.513278 (rank : 12) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P21397, Q16426 | Gene names | MAOA | |||
|
Domain Architecture |

|
|||||
| Description | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A). | |||||
|
OXLA_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 13) | NC score | 0.426153 (rank : 13) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O09046, Q3T9N9, Q3U7S6, Q3V0K2, Q6Y632, Q6YBV6, Q6YDI8, Q8R2G8, Q9CXK7 | Gene names | Il4i1, Fig1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (mFIG1). | |||||
|
OXLA_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 14) | NC score | 0.338107 (rank : 14) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96RQ9, Q4GZN1, Q4GZN2, Q6P2Q3, Q8TEM5, Q96RQ8 | Gene names | IL4I1, FIG1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (hFIG1). | |||||
|
FMO3_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 15) | NC score | 0.107768 (rank : 18) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P97501 | Gene names | Fmo3 | |||
|
Domain Architecture |
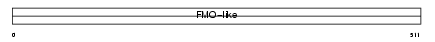
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 3 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 3) (FMO 3) (Dimethylaniline oxidase 3). | |||||
|
ZCPW1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 16) | NC score | 0.080451 (rank : 21) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
CJ033_HUMAN
|
||||||
| θ value | 0.163984 (rank : 17) | NC score | 0.136207 (rank : 17) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N2H3, Q5TAA9, Q9BRQ1 | Gene names | C10orf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 (EC 1.-.-.-). | |||||
|
CJ033_MOUSE
|
||||||
| θ value | 0.163984 (rank : 18) | NC score | 0.137417 (rank : 16) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3U4I7, Q8CAN2, Q9D630 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 homolog (EC 1.-.-.-). | |||||
|
FMO4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.070312 (rank : 25) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P31512 | Gene names | FMO4, FMO2 | |||
|
Domain Architecture |
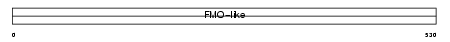
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 4 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 4) (FMO 4) (Dimethylaniline oxidase 4). | |||||
|
MICA2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.063328 (rank : 35) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94851, Q7Z3A8 | Gene names | MICAL2, KIAA0750 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
MICA3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.063251 (rank : 36) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7RTP6, Q6ICK4, Q9P2I3 | Gene names | MICAL3, KIAA1364 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-3. | |||||
|
MICA3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.064617 (rank : 32) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CJ19, Q80TE8, Q8BXB1 | Gene names | Mical3, Kiaa1364 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-3. | |||||
|
MICA2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.064141 (rank : 33) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BML1, Q3UPU6 | Gene names | Mical2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
ERG1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.069695 (rank : 28) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14534, Q9UEK6 | Gene names | SQLE, ERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Squalene monooxygenase (EC 1.14.99.7) (Squalene epoxidase) (SE). | |||||
|
FMO3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.085147 (rank : 20) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P31513, Q14854, Q8N5N5 | Gene names | FMO3 | |||
|
Domain Architecture |
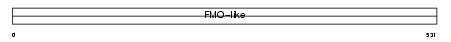
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 3 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 3) (FMO 3) (Dimethylaniline oxidase 3) (FMO form 2) (FMO II). | |||||
|
ETFD_HUMAN
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.059880 (rank : 38) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q16134, Q7Z347 | Gene names | ETFDH | |||
|
Domain Architecture |

|
|||||
| Description | Electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial precursor (EC 1.5.5.1) (ETF-QO) (ETF-ubiquinone oxidoreductase) (ETF dehydrogenase) (Electron-transferring- flavoprotein dehydrogenase). | |||||
|
JAG2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.009600 (rank : 52) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.015315 (rank : 49) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
PPOX_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.173517 (rank : 15) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P50336 | Gene names | PPOX | |||
|
Domain Architecture |
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
ZCPW1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.068858 (rank : 30) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
SMRC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.067491 (rank : 31) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
SMRC1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.070174 (rank : 26) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.073825 (rank : 23) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.073379 (rank : 24) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
DLDH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.052858 (rank : 39) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P09622, Q14131, Q14167 | Gene names | DLD, GCSL, LAD, PHE3 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase) (Glycine cleavage system L protein). | |||||
|
DLDH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.051956 (rank : 42) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08749 | Gene names | Dld | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase). | |||||
|
ETFD_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.037314 (rank : 44) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q921G7, Q3U7K2, Q8BK82, Q9DCT9 | Gene names | Etfdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial precursor (EC 1.5.5.1) (ETF-QO) (ETF-ubiquinone oxidoreductase) (ETF dehydrogenase) (Electron-transferring- flavoprotein dehydrogenase). | |||||
|
FMO2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.069789 (rank : 27) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K2I3, Q9QZF7 | Gene names | Fmo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 2 (EC 1.14.13.8) (Pulmonary flavin-containing monooxygenase 2) (FMO 2) (Dimethylaniline oxidase 2). | |||||
|
MICA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.052564 (rank : 40) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VDP3 | Gene names | Mical1, Mical, Nical | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
DHE4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.029015 (rank : 45) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49448, Q9UDQ4 | Gene names | GLUD2, GLUDP1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate dehydrogenase 2, mitochondrial precursor (EC 1.4.1.3) (GDH). | |||||
|
FMO1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.076051 (rank : 22) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q01740, Q5QPT2, Q9UJC2 | Gene names | FMO1 | |||
|
Domain Architecture |
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 1 (EC 1.14.13.8) (Fetal hepatic flavin-containing monooxygenase 1) (FMO 1) (Dimethylaniline oxidase 1). | |||||
|
FMO6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.069357 (rank : 29) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60774 | Gene names | FMO6 | |||
|
Domain Architecture |
|
|||||
| Description | Putative dimethylaniline monooxygenase [N-oxide-forming] 6 (EC 1.14.13.8) (Flavin-containing monooxygenase 6) (FMO 6) (Dimethylaniline oxidase 6). | |||||
|
FMO2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.063724 (rank : 34) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99518, Q5EBX4, Q9BRX1 | Gene names | FMO2 | |||
|
Domain Architecture |
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 2 (EC 1.14.13.8) (Pulmonary flavin-containing monooxygenase 2) (FMO 2) (Dimethylaniline oxidase 2) (FMO 1B1). | |||||
|
TRXR2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.026424 (rank : 46) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JLT4, Q91YX4, Q9JHA7, Q9JMH5 | Gene names | Txnrd2, Trxr2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 2, mitochondrial precursor (EC 1.8.1.9) (TR3). | |||||
|
AMPO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.010581 (rank : 51) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N6M6, Q5T9B1, Q5T9B3, Q5T9B4, Q8WUL6, Q96M23, Q96SS1 | Gene names | ONPEP, C9orf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aminopeptidase O (EC 3.4.11.-) (AP-O). | |||||
|
DHE3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.024600 (rank : 47) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00367 | Gene names | GLUD1, GLUD | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate dehydrogenase 1, mitochondrial precursor (EC 1.4.1.3) (GDH). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.002177 (rank : 55) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
K0256_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.012032 (rank : 50) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q93073, Q8N767 | Gene names | KIAA0256 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0256. | |||||
|
UCK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.022117 (rank : 48) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52623 | Gene names | Uck1, Umpk | |||
|
Domain Architecture |
|
|||||
| Description | Uridine-cytidine kinase 1 (EC 2.7.1.48) (UCK 1) (Uridine monophosphokinase 1) (Cytidine monophosphokinase 1). | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.008924 (rank : 53) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
FAN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.003180 (rank : 54) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35242 | Gene names | Nsmaf, Fan | |||
|
Domain Architecture |

|
|||||
| Description | Protein FAN (Factor associated with N-SMase activation) (Factor associated with neutral sphingomyelinase activation). | |||||
|
FMO4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.050762 (rank : 43) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHG0 | Gene names | Fmo4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 4 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 4) (FMO 4) (Dimethylaniline oxidase 4). | |||||
|
LAMA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.001920 (rank : 56) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
FMO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.060587 (rank : 37) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P50285 | Gene names | Fmo1, Fmo-1 | |||
|
Domain Architecture |
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 1 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 1) (FMO 1) (Dimethylaniline oxidase 1). | |||||
|
PCYOX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.052324 (rank : 41) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CQF9, Q3UHV6, Q69ZW0, Q8BZX1 | Gene names | Pcyox1, Kiaa0908 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prenylcysteine oxidase precursor (EC 1.8.3.5). | |||||
|
PPOX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.099807 (rank : 19) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51175, P97344, Q99M34 | Gene names | Ppox | |||
|
Domain Architecture |
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
AOF1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8NB78, Q5TGV3, Q6AI15, Q6ZUU4, Q8N258, Q96EL7 | Gene names | AOF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
AOF1_MOUSE
|
||||||
| NC score | 0.997324 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CIG3, Q8C5C4, Q8CEC1 | Gene names | Aof1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
LSD1_MOUSE
|
||||||
| NC score | 0.799463 (rank : 3) | θ value | 1.46268e-50 (rank : 4) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6ZQ88, Q6PB53, Q8VEA1 | Gene names | Aof2, Kiaa0601, Lsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
LSD1_HUMAN
|
||||||
| NC score | 0.764986 (rank : 4) | θ value | 1.46268e-50 (rank : 3) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
PAOX_MOUSE
|
||||||
| NC score | 0.759609 (rank : 5) | θ value | 8.3442e-30 (rank : 5) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C0L6, Q8K254 | Gene names | Paox, Pao | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal N1-acetyl-spermine/spermidine oxidase (EC 1.5.3.11) (Polyamine oxidase). | |||||
|
SMOX_HUMAN
|
||||||
| NC score | 0.729835 (rank : 6) | θ value | 1.02001e-27 (rank : 6) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NWM0, Q5TE26, Q5TE27, Q6UY28, Q8IX00, Q96LC3, Q96LC4, Q96QT3, Q9BW38, Q9H6H1, Q9NP51, Q9NPY1, Q9NPY2 | Gene names | SMOX, C20orf16, SMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermine oxidase (EC 1.5.3.-) (Polyamine oxidase 1) (PAO-1) (PAOh1). | |||||
|
SMOX_MOUSE
|
||||||
| NC score | 0.722427 (rank : 7) | θ value | 1.92365e-26 (rank : 7) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99K82, Q70LA3, Q70LA4, Q70LA5, Q70LA7, Q70LA8, Q70LA9, Q70LB0, Q8CJ56, Q8CJ57 | Gene names | Smox, Smo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermine oxidase (EC 1.5.3.-) (Polyamine oxidase 1) (PAO-1) (PAOh1). | |||||
|
PAOX_HUMAN
|
||||||
| NC score | 0.720134 (rank : 8) | θ value | 3.07116e-24 (rank : 8) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6QHF9, Q6QHF5, Q6QHF6, Q6QHF7, Q6QHF8, Q6QHG0, Q6QHG1, Q6QHG2, Q6QHG3, Q6QHG4, Q6QHG5, Q6QHG6, Q86WP9, Q8N555, Q8NCX3 | Gene names | PAOX, PAO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal N1-acetyl-spermine/spermidine oxidase (EC 1.5.3.11) (Polyamine oxidase). | |||||
|
AOFB_HUMAN
|
||||||
| NC score | 0.575645 (rank : 9) | θ value | 3.63628e-17 (rank : 9) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P27338, Q7Z6S2 | Gene names | MAOB | |||
|
Domain Architecture |
|
|||||
| Description | Amine oxidase [flavin-containing] B (EC 1.4.3.4) (Monoamine oxidase type B) (MAO-B). | |||||
|
AOFB_MOUSE
|
||||||
| NC score | 0.553924 (rank : 10) | θ value | 1.058e-16 (rank : 10) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BW75, Q8C0B2 | Gene names | Maob | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amine oxidase [flavin-containing] B (EC 1.4.3.4) (Monoamine oxidase type B) (MAO-B). | |||||
|
AOFA_MOUSE
|
||||||
| NC score | 0.547626 (rank : 11) | θ value | 1.52774e-15 (rank : 11) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q64133, Q8K0Z8 | Gene names | Maoa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A). | |||||
|
AOFA_HUMAN
|
||||||
| NC score | 0.513278 (rank : 12) | θ value | 3.52202e-12 (rank : 12) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P21397, Q16426 | Gene names | MAOA | |||
|
Domain Architecture |

|
|||||
| Description | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A). | |||||
|
OXLA_MOUSE
|
||||||
| NC score | 0.426153 (rank : 13) | θ value | 4.45536e-07 (rank : 13) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O09046, Q3T9N9, Q3U7S6, Q3V0K2, Q6Y632, Q6YBV6, Q6YDI8, Q8R2G8, Q9CXK7 | Gene names | Il4i1, Fig1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (mFIG1). | |||||
|
OXLA_HUMAN
|
||||||
| NC score | 0.338107 (rank : 14) | θ value | 0.00869519 (rank : 14) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96RQ9, Q4GZN1, Q4GZN2, Q6P2Q3, Q8TEM5, Q96RQ8 | Gene names | IL4I1, FIG1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (hFIG1). | |||||
|
PPOX_HUMAN
|
||||||
| NC score | 0.173517 (rank : 15) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P50336 | Gene names | PPOX | |||
|
Domain Architecture |
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
CJ033_MOUSE
|
||||||
| NC score | 0.137417 (rank : 16) | θ value | 0.163984 (rank : 18) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3U4I7, Q8CAN2, Q9D630 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 homolog (EC 1.-.-.-). | |||||
|
CJ033_HUMAN
|
||||||
| NC score | 0.136207 (rank : 17) | θ value | 0.163984 (rank : 17) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N2H3, Q5TAA9, Q9BRQ1 | Gene names | C10orf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 (EC 1.-.-.-). | |||||
|
FMO3_MOUSE
|
||||||
| NC score | 0.107768 (rank : 18) | θ value | 0.0431538 (rank : 15) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P97501 | Gene names | Fmo3 | |||
|
Domain Architecture |
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 3 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 3) (FMO 3) (Dimethylaniline oxidase 3). | |||||
|
PPOX_MOUSE
|
||||||
| NC score | 0.099807 (rank : 19) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51175, P97344, Q99M34 | Gene names | Ppox | |||
|
Domain Architecture |
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
FMO3_HUMAN
|
||||||
| NC score | 0.085147 (rank : 20) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P31513, Q14854, Q8N5N5 | Gene names | FMO3 | |||
|
Domain Architecture |
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 3 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 3) (FMO 3) (Dimethylaniline oxidase 3) (FMO form 2) (FMO II). | |||||
|
ZCPW1_MOUSE
|
||||||
| NC score | 0.080451 (rank : 21) | θ value | 0.0431538 (rank : 16) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
FMO1_HUMAN
|
||||||
| NC score | 0.076051 (rank : 22) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q01740, Q5QPT2, Q9UJC2 | Gene names | FMO1 | |||
|
Domain Architecture |
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 1 (EC 1.14.13.8) (Fetal hepatic flavin-containing monooxygenase 1) (FMO 1) (Dimethylaniline oxidase 1). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.073825 (rank : 23) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.073379 (rank : 24) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
FMO4_HUMAN
|
||||||
| NC score | 0.070312 (rank : 25) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P31512 | Gene names | FMO4, FMO2 | |||
|
Domain Architecture |
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 4 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 4) (FMO 4) (Dimethylaniline oxidase 4). | |||||
|
SMRC1_MOUSE
|
||||||
| NC score | 0.070174 (rank : 26) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
FMO2_MOUSE
|
||||||
| NC score | 0.069789 (rank : 27) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K2I3, Q9QZF7 | Gene names | Fmo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 2 (EC 1.14.13.8) (Pulmonary flavin-containing monooxygenase 2) (FMO 2) (Dimethylaniline oxidase 2). | |||||
|
ERG1_HUMAN
|
||||||
| NC score | 0.069695 (rank : 28) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14534, Q9UEK6 | Gene names | SQLE, ERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Squalene monooxygenase (EC 1.14.99.7) (Squalene epoxidase) (SE). | |||||
|
FMO6_HUMAN
|
||||||
| NC score | 0.069357 (rank : 29) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60774 | Gene names | FMO6 | |||
|
Domain Architecture |
|
|||||
| Description | Putative dimethylaniline monooxygenase [N-oxide-forming] 6 (EC 1.14.13.8) (Flavin-containing monooxygenase 6) (FMO 6) (Dimethylaniline oxidase 6). | |||||
|
ZCPW1_HUMAN
|
||||||
| NC score | 0.068858 (rank : 30) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
SMRC1_HUMAN
|
||||||
| NC score | 0.067491 (rank : 31) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
MICA3_MOUSE
|
||||||
| NC score | 0.064617 (rank : 32) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CJ19, Q80TE8, Q8BXB1 | Gene names | Mical3, Kiaa1364 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-3. | |||||
|
MICA2_MOUSE
|
||||||
| NC score | 0.064141 (rank : 33) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BML1, Q3UPU6 | Gene names | Mical2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
FMO2_HUMAN
|
||||||
| NC score | 0.063724 (rank : 34) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99518, Q5EBX4, Q9BRX1 | Gene names | FMO2 | |||
|
Domain Architecture |
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 2 (EC 1.14.13.8) (Pulmonary flavin-containing monooxygenase 2) (FMO 2) (Dimethylaniline oxidase 2) (FMO 1B1). | |||||
|
MICA2_HUMAN
|
||||||
| NC score | 0.063328 (rank : 35) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94851, Q7Z3A8 | Gene names | MICAL2, KIAA0750 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
MICA3_HUMAN
|
||||||
| NC score | 0.063251 (rank : 36) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7RTP6, Q6ICK4, Q9P2I3 | Gene names | MICAL3, KIAA1364 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-3. | |||||
|
FMO1_MOUSE
|
||||||
| NC score | 0.060587 (rank : 37) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P50285 | Gene names | Fmo1, Fmo-1 | |||
|
Domain Architecture |
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 1 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 1) (FMO 1) (Dimethylaniline oxidase 1). | |||||
|
ETFD_HUMAN
|
||||||
| NC score | 0.059880 (rank : 38) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q16134, Q7Z347 | Gene names | ETFDH | |||
|
Domain Architecture |

|
|||||
| Description | Electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial precursor (EC 1.5.5.1) (ETF-QO) (ETF-ubiquinone oxidoreductase) (ETF dehydrogenase) (Electron-transferring- flavoprotein dehydrogenase). | |||||
|
DLDH_HUMAN
|
||||||
| NC score | 0.052858 (rank : 39) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P09622, Q14131, Q14167 | Gene names | DLD, GCSL, LAD, PHE3 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase) (Glycine cleavage system L protein). | |||||
|
MICA1_MOUSE
|
||||||
| NC score | 0.052564 (rank : 40) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VDP3 | Gene names | Mical1, Mical, Nical | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
PCYOX_MOUSE
|
||||||
| NC score | 0.052324 (rank : 41) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CQF9, Q3UHV6, Q69ZW0, Q8BZX1 | Gene names | Pcyox1, Kiaa0908 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prenylcysteine oxidase precursor (EC 1.8.3.5). | |||||
|
DLDH_MOUSE
|
||||||
| NC score | 0.051956 (rank : 42) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08749 | Gene names | Dld | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase). | |||||
|
FMO4_MOUSE
|
||||||
| NC score | 0.050762 (rank : 43) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHG0 | Gene names | Fmo4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 4 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 4) (FMO 4) (Dimethylaniline oxidase 4). | |||||
|
ETFD_MOUSE
|
||||||
| NC score | 0.037314 (rank : 44) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q921G7, Q3U7K2, Q8BK82, Q9DCT9 | Gene names | Etfdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial precursor (EC 1.5.5.1) (ETF-QO) (ETF-ubiquinone oxidoreductase) (ETF dehydrogenase) (Electron-transferring- flavoprotein dehydrogenase). | |||||
|
DHE4_HUMAN
|
||||||
| NC score | 0.029015 (rank : 45) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49448, Q9UDQ4 | Gene names | GLUD2, GLUDP1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate dehydrogenase 2, mitochondrial precursor (EC 1.4.1.3) (GDH). | |||||
|
TRXR2_MOUSE
|
||||||
| NC score | 0.026424 (rank : 46) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JLT4, Q91YX4, Q9JHA7, Q9JMH5 | Gene names | Txnrd2, Trxr2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 2, mitochondrial precursor (EC 1.8.1.9) (TR3). | |||||
|
DHE3_HUMAN
|
||||||
| NC score | 0.024600 (rank : 47) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00367 | Gene names | GLUD1, GLUD | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate dehydrogenase 1, mitochondrial precursor (EC 1.4.1.3) (GDH). | |||||
|
UCK1_MOUSE
|
||||||
| NC score | 0.022117 (rank : 48) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52623 | Gene names | Uck1, Umpk | |||
|
Domain Architecture |

|
|||||
| Description | Uridine-cytidine kinase 1 (EC 2.7.1.48) (UCK 1) (Uridine monophosphokinase 1) (Cytidine monophosphokinase 1). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.015315 (rank : 49) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
K0256_HUMAN
|
||||||
| NC score | 0.012032 (rank : 50) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q93073, Q8N767 | Gene names | KIAA0256 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0256. | |||||
|
AMPO_HUMAN
|
||||||
| NC score | 0.010581 (rank : 51) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N6M6, Q5T9B1, Q5T9B3, Q5T9B4, Q8WUL6, Q96M23, Q96SS1 | Gene names | ONPEP, C9orf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aminopeptidase O (EC 3.4.11.-) (AP-O). | |||||
|
JAG2_MOUSE
|
||||||
| NC score | 0.009600 (rank : 52) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.008924 (rank : 53) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
FAN_MOUSE
|
||||||
| NC score | 0.003180 (rank : 54) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35242 | Gene names | Nsmaf, Fan | |||
|
Domain Architecture |

|
|||||
| Description | Protein FAN (Factor associated with N-SMase activation) (Factor associated with neutral sphingomyelinase activation). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.002177 (rank : 55) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
LAMA1_MOUSE
|
||||||
| NC score | 0.001920 (rank : 56) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||