Please be patient as the page loads
|
PPOX_HUMAN
|
||||||
| SwissProt Accessions | P50336 | Gene names | PPOX | |||
|
Domain Architecture |
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PPOX_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P50336 | Gene names | PPOX | |||
|
Domain Architecture |
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
PPOX_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.987433 (rank : 2) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P51175, P97344, Q99M34 | Gene names | Ppox | |||
|
Domain Architecture |
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
AOFB_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 3) | NC score | 0.263820 (rank : 4) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BW75, Q8C0B2 | Gene names | Maob | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amine oxidase [flavin-containing] B (EC 1.4.3.4) (Monoamine oxidase type B) (MAO-B). | |||||
|
AOFB_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 4) | NC score | 0.276286 (rank : 3) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P27338, Q7Z6S2 | Gene names | MAOB | |||
|
Domain Architecture |
|
|||||
| Description | Amine oxidase [flavin-containing] B (EC 1.4.3.4) (Monoamine oxidase type B) (MAO-B). | |||||
|
AOFA_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 5) | NC score | 0.259996 (rank : 5) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64133, Q8K0Z8 | Gene names | Maoa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A). | |||||
|
PAOX_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 6) | NC score | 0.221378 (rank : 7) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6QHF9, Q6QHF5, Q6QHF6, Q6QHF7, Q6QHF8, Q6QHG0, Q6QHG1, Q6QHG2, Q6QHG3, Q6QHG4, Q6QHG5, Q6QHG6, Q86WP9, Q8N555, Q8NCX3 | Gene names | PAOX, PAO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal N1-acetyl-spermine/spermidine oxidase (EC 1.5.3.11) (Polyamine oxidase). | |||||
|
AOFA_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 7) | NC score | 0.237662 (rank : 6) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P21397, Q16426 | Gene names | MAOA | |||
|
Domain Architecture |

|
|||||
| Description | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A). | |||||
|
AOF1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 8) | NC score | 0.194930 (rank : 9) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CIG3, Q8C5C4, Q8CEC1 | Gene names | Aof1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
PAOX_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 9) | NC score | 0.221049 (rank : 8) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C0L6, Q8K254 | Gene names | Paox, Pao | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal N1-acetyl-spermine/spermidine oxidase (EC 1.5.3.11) (Polyamine oxidase). | |||||
|
CJ033_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 10) | NC score | 0.118660 (rank : 17) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3U4I7, Q8CAN2, Q9D630 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 homolog (EC 1.-.-.-). | |||||
|
OXLA_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 11) | NC score | 0.184382 (rank : 10) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96RQ9, Q4GZN1, Q4GZN2, Q6P2Q3, Q8TEM5, Q96RQ8 | Gene names | IL4I1, FIG1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (hFIG1). | |||||
|
HDAC6_MOUSE
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.032453 (rank : 24) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2V5 | Gene names | Hdac6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6) (Histone deacetylase mHDA2). | |||||
|
OXLA_MOUSE
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.181077 (rank : 11) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O09046, Q3T9N9, Q3U7S6, Q3V0K2, Q6Y632, Q6YBV6, Q6YDI8, Q8R2G8, Q9CXK7 | Gene names | Il4i1, Fig1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (mFIG1). | |||||
|
RETST_HUMAN
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.090024 (rank : 18) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6NUM9, Q53R95, Q53SA9, Q6UX05, Q8N2H5, Q96FA4, Q9NXE5 | Gene names | RETSAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
SMOX_HUMAN
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.174476 (rank : 13) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NWM0, Q5TE26, Q5TE27, Q6UY28, Q8IX00, Q96LC3, Q96LC4, Q96QT3, Q9BW38, Q9H6H1, Q9NP51, Q9NPY1, Q9NPY2 | Gene names | SMOX, C20orf16, SMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermine oxidase (EC 1.5.3.-) (Polyamine oxidase 1) (PAO-1) (PAOh1). | |||||
|
SMOX_MOUSE
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.179092 (rank : 12) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99K82, Q70LA3, Q70LA4, Q70LA5, Q70LA7, Q70LA8, Q70LA9, Q70LB0, Q8CJ56, Q8CJ57 | Gene names | Smox, Smo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermine oxidase (EC 1.5.3.-) (Polyamine oxidase 1) (PAO-1) (PAOh1). | |||||
|
LSD1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.144324 (rank : 16) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
LSD1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.151246 (rank : 15) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6ZQ88, Q6PB53, Q8VEA1 | Gene names | Aof2, Kiaa0601, Lsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
SON_MOUSE
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.032435 (rank : 25) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
TYK2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.008969 (rank : 33) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R117, O88431, Q8VE41 | Gene names | Tyk2 | |||
|
Domain Architecture |

|
|||||
| Description | Non-receptor tyrosine-protein kinase TYK2 (EC 2.7.10.2). | |||||
|
AOF1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.173517 (rank : 14) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NB78, Q5TGV3, Q6AI15, Q6ZUU4, Q8N258, Q96EL7 | Gene names | AOF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
URP2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.026197 (rank : 26) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K1B8 | Gene names | Urp2, Kind3 | |||
|
Domain Architecture |

|
|||||
| Description | Unc-112-related protein 2 (Kindlin-3). | |||||
|
M2GD_HUMAN
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.042737 (rank : 22) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UI17 | Gene names | DMGDH | |||
|
Domain Architecture |

|
|||||
| Description | Dimethylglycine dehydrogenase, mitochondrial precursor (EC 1.5.99.2) (ME2GLYDH). | |||||
|
SPR1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.014651 (rank : 31) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15743, Q13334, Q6IX34 | Gene names | GPR68, OGR1 | |||
|
Domain Architecture |
|
|||||
| Description | Sphingosylphosphorylcholine receptor (Ovarian cancer G-protein coupled receptor 1) (OGR-1) (G-protein coupled receptor 68) (GPR12A). | |||||
|
FAS_MOUSE
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.024522 (rank : 27) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19096, Q6PB72, Q8C4Z0, Q9EQR0 | Gene names | Fasn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
M2GD_MOUSE
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.040584 (rank : 23) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBT9, Q8R1S7 | Gene names | Dmgdh | |||
|
Domain Architecture |

|
|||||
| Description | Dimethylglycine dehydrogenase, mitochondrial precursor (EC 1.5.99.2) (ME2GLYDH). | |||||
|
SPR1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.013957 (rank : 32) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BFQ3, Q3U1L3, Q80T34 | Gene names | Gpr68 | |||
|
Domain Architecture |
|
|||||
| Description | Sphingosylphosphorylcholine receptor (G-protein coupled receptor 68). | |||||
|
RETST_MOUSE
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.079613 (rank : 19) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q64FW2, Q3UNN9, Q78JX8, Q8VHE7, Q9CW85 | Gene names | Retsat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
PRRT3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.022580 (rank : 28) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SON_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.017158 (rank : 29) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
PCYOX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.044018 (rank : 21) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CQF9, Q3UHV6, Q69ZW0, Q8BZX1 | Gene names | Pcyox1, Kiaa0908 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prenylcysteine oxidase precursor (EC 1.8.3.5). | |||||
|
IQCB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.014766 (rank : 30) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15051, Q5DKQ7, Q8NI79, Q9BS08 | Gene names | IQCB1, KIAA0036, NPHP5 | |||
|
Domain Architecture |

|
|||||
| Description | IQ calmodulin-binding motif-containing protein 1 (Nephrocystin-5). | |||||
|
MBL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.007510 (rank : 35) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41317 | Gene names | Mbl2 | |||
|
Domain Architecture |
|
|||||
| Description | Mannose-binding protein C precursor (MBP-C) (Mannan-binding protein) (RA-reactive factor P28A subunit) (RARF/P28A). | |||||
|
TF7L2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.008132 (rank : 34) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQB0, O00185, Q9NQB1, Q9NQB2, Q9NQB3, Q9NQB4, Q9NQB5, Q9NQB6, Q9NQB7, Q9ULC2 | Gene names | TCF7L2, TCF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (hTCF-4). | |||||
|
ZN628_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | -0.000606 (rank : 36) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5EBL2, Q86X34 | Gene names | ZNF628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 628. | |||||
|
CJ033_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.066123 (rank : 20) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N2H3, Q5TAA9, Q9BRQ1 | Gene names | C10orf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 (EC 1.-.-.-). | |||||
|
PPOX_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P50336 | Gene names | PPOX | |||
|
Domain Architecture |
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
PPOX_MOUSE
|
||||||
| NC score | 0.987433 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P51175, P97344, Q99M34 | Gene names | Ppox | |||
|
Domain Architecture |
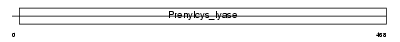
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
AOFB_HUMAN
|
||||||
| NC score | 0.276286 (rank : 3) | θ value | 0.00228821 (rank : 4) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P27338, Q7Z6S2 | Gene names | MAOB | |||
|
Domain Architecture |
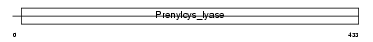
|
|||||
| Description | Amine oxidase [flavin-containing] B (EC 1.4.3.4) (Monoamine oxidase type B) (MAO-B). | |||||
|
AOFB_MOUSE
|
||||||
| NC score | 0.263820 (rank : 4) | θ value | 9.29e-05 (rank : 3) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BW75, Q8C0B2 | Gene names | Maob | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amine oxidase [flavin-containing] B (EC 1.4.3.4) (Monoamine oxidase type B) (MAO-B). | |||||
|
AOFA_MOUSE
|
||||||
| NC score | 0.259996 (rank : 5) | θ value | 0.00390308 (rank : 5) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64133, Q8K0Z8 | Gene names | Maoa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A). | |||||
|
AOFA_HUMAN
|
||||||
| NC score | 0.237662 (rank : 6) | θ value | 0.0113563 (rank : 7) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P21397, Q16426 | Gene names | MAOA | |||
|
Domain Architecture |

|
|||||
| Description | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A). | |||||
|
PAOX_HUMAN
|
||||||
| NC score | 0.221378 (rank : 7) | θ value | 0.00509761 (rank : 6) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6QHF9, Q6QHF5, Q6QHF6, Q6QHF7, Q6QHF8, Q6QHG0, Q6QHG1, Q6QHG2, Q6QHG3, Q6QHG4, Q6QHG5, Q6QHG6, Q86WP9, Q8N555, Q8NCX3 | Gene names | PAOX, PAO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal N1-acetyl-spermine/spermidine oxidase (EC 1.5.3.11) (Polyamine oxidase). | |||||
|
PAOX_MOUSE
|
||||||
| NC score | 0.221049 (rank : 8) | θ value | 0.0431538 (rank : 9) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C0L6, Q8K254 | Gene names | Paox, Pao | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal N1-acetyl-spermine/spermidine oxidase (EC 1.5.3.11) (Polyamine oxidase). | |||||
|
AOF1_MOUSE
|
||||||
| NC score | 0.194930 (rank : 9) | θ value | 0.0330416 (rank : 8) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CIG3, Q8C5C4, Q8CEC1 | Gene names | Aof1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
OXLA_HUMAN
|
||||||
| NC score | 0.184382 (rank : 10) | θ value | 0.0961366 (rank : 11) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96RQ9, Q4GZN1, Q4GZN2, Q6P2Q3, Q8TEM5, Q96RQ8 | Gene names | IL4I1, FIG1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (hFIG1). | |||||
|
OXLA_MOUSE
|
||||||
| NC score | 0.181077 (rank : 11) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O09046, Q3T9N9, Q3U7S6, Q3V0K2, Q6Y632, Q6YBV6, Q6YDI8, Q8R2G8, Q9CXK7 | Gene names | Il4i1, Fig1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (mFIG1). | |||||
|
SMOX_MOUSE
|
||||||
| NC score | 0.179092 (rank : 12) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99K82, Q70LA3, Q70LA4, Q70LA5, Q70LA7, Q70LA8, Q70LA9, Q70LB0, Q8CJ56, Q8CJ57 | Gene names | Smox, Smo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermine oxidase (EC 1.5.3.-) (Polyamine oxidase 1) (PAO-1) (PAOh1). | |||||
|
SMOX_HUMAN
|
||||||
| NC score | 0.174476 (rank : 13) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NWM0, Q5TE26, Q5TE27, Q6UY28, Q8IX00, Q96LC3, Q96LC4, Q96QT3, Q9BW38, Q9H6H1, Q9NP51, Q9NPY1, Q9NPY2 | Gene names | SMOX, C20orf16, SMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermine oxidase (EC 1.5.3.-) (Polyamine oxidase 1) (PAO-1) (PAOh1). | |||||
|
AOF1_HUMAN
|
||||||
| NC score | 0.173517 (rank : 14) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NB78, Q5TGV3, Q6AI15, Q6ZUU4, Q8N258, Q96EL7 | Gene names | AOF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
LSD1_MOUSE
|
||||||
| NC score | 0.151246 (rank : 15) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6ZQ88, Q6PB53, Q8VEA1 | Gene names | Aof2, Kiaa0601, Lsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
LSD1_HUMAN
|
||||||
| NC score | 0.144324 (rank : 16) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
CJ033_MOUSE
|
||||||
| NC score | 0.118660 (rank : 17) | θ value | 0.0563607 (rank : 10) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3U4I7, Q8CAN2, Q9D630 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 homolog (EC 1.-.-.-). | |||||
|
RETST_HUMAN
|
||||||
| NC score | 0.090024 (rank : 18) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6NUM9, Q53R95, Q53SA9, Q6UX05, Q8N2H5, Q96FA4, Q9NXE5 | Gene names | RETSAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
RETST_MOUSE
|
||||||
| NC score | 0.079613 (rank : 19) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q64FW2, Q3UNN9, Q78JX8, Q8VHE7, Q9CW85 | Gene names | Retsat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
CJ033_HUMAN
|
||||||
| NC score | 0.066123 (rank : 20) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N2H3, Q5TAA9, Q9BRQ1 | Gene names | C10orf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 (EC 1.-.-.-). | |||||
|
PCYOX_MOUSE
|
||||||
| NC score | 0.044018 (rank : 21) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CQF9, Q3UHV6, Q69ZW0, Q8BZX1 | Gene names | Pcyox1, Kiaa0908 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prenylcysteine oxidase precursor (EC 1.8.3.5). | |||||
|
M2GD_HUMAN
|
||||||
| NC score | 0.042737 (rank : 22) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UI17 | Gene names | DMGDH | |||
|
Domain Architecture |

|
|||||
| Description | Dimethylglycine dehydrogenase, mitochondrial precursor (EC 1.5.99.2) (ME2GLYDH). | |||||
|
M2GD_MOUSE
|
||||||
| NC score | 0.040584 (rank : 23) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBT9, Q8R1S7 | Gene names | Dmgdh | |||
|
Domain Architecture |

|
|||||
| Description | Dimethylglycine dehydrogenase, mitochondrial precursor (EC 1.5.99.2) (ME2GLYDH). | |||||
|
HDAC6_MOUSE
|
||||||
| NC score | 0.032453 (rank : 24) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2V5 | Gene names | Hdac6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6) (Histone deacetylase mHDA2). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.032435 (rank : 25) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
URP2_MOUSE
|
||||||
| NC score | 0.026197 (rank : 26) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K1B8 | Gene names | Urp2, Kind3 | |||
|
Domain Architecture |

|
|||||
| Description | Unc-112-related protein 2 (Kindlin-3). | |||||
|
FAS_MOUSE
|
||||||
| NC score | 0.024522 (rank : 27) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19096, Q6PB72, Q8C4Z0, Q9EQR0 | Gene names | Fasn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
PRRT3_MOUSE
|
||||||
| NC score | 0.022580 (rank : 28) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.017158 (rank : 29) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
IQCB1_HUMAN
|
||||||
| NC score | 0.014766 (rank : 30) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15051, Q5DKQ7, Q8NI79, Q9BS08 | Gene names | IQCB1, KIAA0036, NPHP5 | |||
|
Domain Architecture |

|
|||||
| Description | IQ calmodulin-binding motif-containing protein 1 (Nephrocystin-5). | |||||
|
SPR1_HUMAN
|
||||||
| NC score | 0.014651 (rank : 31) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15743, Q13334, Q6IX34 | Gene names | GPR68, OGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosylphosphorylcholine receptor (Ovarian cancer G-protein coupled receptor 1) (OGR-1) (G-protein coupled receptor 68) (GPR12A). | |||||
|
SPR1_MOUSE
|
||||||
| NC score | 0.013957 (rank : 32) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BFQ3, Q3U1L3, Q80T34 | Gene names | Gpr68 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosylphosphorylcholine receptor (G-protein coupled receptor 68). | |||||
|
TYK2_MOUSE
|
||||||
| NC score | 0.008969 (rank : 33) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R117, O88431, Q8VE41 | Gene names | Tyk2 | |||
|
Domain Architecture |

|
|||||
| Description | Non-receptor tyrosine-protein kinase TYK2 (EC 2.7.10.2). | |||||
|
TF7L2_HUMAN
|
||||||
| NC score | 0.008132 (rank : 34) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQB0, O00185, Q9NQB1, Q9NQB2, Q9NQB3, Q9NQB4, Q9NQB5, Q9NQB6, Q9NQB7, Q9ULC2 | Gene names | TCF7L2, TCF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (hTCF-4). | |||||
|
MBL2_MOUSE
|
||||||
| NC score | 0.007510 (rank : 35) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41317 | Gene names | Mbl2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannose-binding protein C precursor (MBP-C) (Mannan-binding protein) (RA-reactive factor P28A subunit) (RARF/P28A). | |||||
|
ZN628_HUMAN
|
||||||
| NC score | -0.000606 (rank : 36) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5EBL2, Q86X34 | Gene names | ZNF628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 628. | |||||