Please be patient as the page loads
|
AOFA_MOUSE
|
||||||
| SwissProt Accessions | Q64133, Q8K0Z8 | Gene names | Maoa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AOFA_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.987545 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P21397, Q16426 | Gene names | MAOA | |||
|
Domain Architecture |

|
|||||
| Description | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A). | |||||
|
AOFA_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q64133, Q8K0Z8 | Gene names | Maoa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A). | |||||
|
AOFB_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.989564 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P27338, Q7Z6S2 | Gene names | MAOB | |||
|
Domain Architecture |
|
|||||
| Description | Amine oxidase [flavin-containing] B (EC 1.4.3.4) (Monoamine oxidase type B) (MAO-B). | |||||
|
AOFB_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.987861 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BW75, Q8C0B2 | Gene names | Maob | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amine oxidase [flavin-containing] B (EC 1.4.3.4) (Monoamine oxidase type B) (MAO-B). | |||||
|
AOF1_HUMAN
|
||||||
| θ value | 1.52774e-15 (rank : 5) | NC score | 0.547626 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NB78, Q5TGV3, Q6AI15, Q6ZUU4, Q8N258, Q96EL7 | Gene names | AOF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
AOF1_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 6) | NC score | 0.543833 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CIG3, Q8C5C4, Q8CEC1 | Gene names | Aof1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
OXLA_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 7) | NC score | 0.499876 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O09046, Q3T9N9, Q3U7S6, Q3V0K2, Q6Y632, Q6YBV6, Q6YDI8, Q8R2G8, Q9CXK7 | Gene names | Il4i1, Fig1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (mFIG1). | |||||
|
OXLA_HUMAN
|
||||||
| θ value | 7.34386e-10 (rank : 8) | NC score | 0.470552 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96RQ9, Q4GZN1, Q4GZN2, Q6P2Q3, Q8TEM5, Q96RQ8 | Gene names | IL4I1, FIG1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (hFIG1). | |||||
|
LSD1_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 9) | NC score | 0.403246 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
LSD1_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 10) | NC score | 0.420796 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZQ88, Q6PB53, Q8VEA1 | Gene names | Aof2, Kiaa0601, Lsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
SMOX_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 11) | NC score | 0.330025 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NWM0, Q5TE26, Q5TE27, Q6UY28, Q8IX00, Q96LC3, Q96LC4, Q96QT3, Q9BW38, Q9H6H1, Q9NP51, Q9NPY1, Q9NPY2 | Gene names | SMOX, C20orf16, SMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermine oxidase (EC 1.5.3.-) (Polyamine oxidase 1) (PAO-1) (PAOh1). | |||||
|
PAOX_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 12) | NC score | 0.415194 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C0L6, Q8K254 | Gene names | Paox, Pao | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal N1-acetyl-spermine/spermidine oxidase (EC 1.5.3.11) (Polyamine oxidase). | |||||
|
RETST_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 13) | NC score | 0.182123 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6NUM9, Q53R95, Q53SA9, Q6UX05, Q8N2H5, Q96FA4, Q9NXE5 | Gene names | RETSAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
SMOX_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 14) | NC score | 0.320009 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99K82, Q70LA3, Q70LA4, Q70LA5, Q70LA7, Q70LA8, Q70LA9, Q70LB0, Q8CJ56, Q8CJ57 | Gene names | Smox, Smo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermine oxidase (EC 1.5.3.-) (Polyamine oxidase 1) (PAO-1) (PAOh1). | |||||
|
RETST_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 15) | NC score | 0.173162 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64FW2, Q3UNN9, Q78JX8, Q8VHE7, Q9CW85 | Gene names | Retsat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
PPOX_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 16) | NC score | 0.259996 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P50336 | Gene names | PPOX | |||
|
Domain Architecture |
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
PAOX_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 17) | NC score | 0.333806 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6QHF9, Q6QHF5, Q6QHF6, Q6QHF7, Q6QHF8, Q6QHG0, Q6QHG1, Q6QHG2, Q6QHG3, Q6QHG4, Q6QHG5, Q6QHG6, Q86WP9, Q8N555, Q8NCX3 | Gene names | PAOX, PAO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal N1-acetyl-spermine/spermidine oxidase (EC 1.5.3.11) (Polyamine oxidase). | |||||
|
PCYOX_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 18) | NC score | 0.105419 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHG3, O94982, Q8N4N5, Q96QM8 | Gene names | PCYOX1, KIAA0908, PCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Prenylcysteine oxidase precursor (EC 1.8.3.5) (PCL1). | |||||
|
PPOX_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 19) | NC score | 0.180940 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P51175, P97344, Q99M34 | Gene names | Ppox | |||
|
Domain Architecture |
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
CJ033_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 20) | NC score | 0.175553 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N2H3, Q5TAA9, Q9BRQ1 | Gene names | C10orf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 (EC 1.-.-.-). | |||||
|
CJ033_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 21) | NC score | 0.184408 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3U4I7, Q8CAN2, Q9D630 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 homolog (EC 1.-.-.-). | |||||
|
TRXR1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 22) | NC score | 0.053479 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JMH6 | Gene names | Txnrd1, Trxr1 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 1, cytoplasmic (EC 1.8.1.9) (TR) (TR1). | |||||
|
TRXR1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.058439 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16881, Q76P53, Q7LA96, Q9UH79 | Gene names | TXNRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 1, cytoplasmic precursor (EC 1.8.1.9) (TR) (TR1). | |||||
|
FMO5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.024604 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49326 | Gene names | FMO5 | |||
|
Domain Architecture |
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 5 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 5) (FMO 5) (Dimethylaniline oxidase 5). | |||||
|
GSHR_MOUSE
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.047266 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P47791, Q7TNC2, Q8BN97, Q8C9Z6 | Gene names | Gsr, Gr1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione reductase, mitochondrial precursor (EC 1.8.1.7) (GR) (GRase). | |||||
|
L2HDH_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.042824 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H9P8, Q9BRR1 | Gene names | L2HGDH, C14orf160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L-2-hydroxyglutarate dehydrogenase, mitochondrial precursor (EC 1.1.99.2) (Duranin). | |||||
|
L2HDH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.043678 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91YP0, Q3TH61, Q3U7Z0, Q3ULY6 | Gene names | L2hgdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L-2-hydroxyglutarate dehydrogenase, mitochondrial precursor (EC 1.1.99.2) (Duranin). | |||||
|
AMID_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.021771 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BRQ8, Q63Z39 | Gene names | AMID, PRG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-inducing factor-like mitochondrion-associated inducer of death (EC 1.-.-.-) (Apoptosis-inducing factor-homologous mitochondrion-associated inducer of death) (p53-responsive gene 3 protein). | |||||
|
DHSA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.022939 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K2B3, Q3UH25 | Gene names | Sdha | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial precursor (EC 1.3.5.1) (Fp) (Flavoprotein subunit of complex II). | |||||
|
GSHR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.043143 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P00390, Q7Z5C9, Q9NP63 | Gene names | GSR, GLUR, GRD1 | |||
|
Domain Architecture |
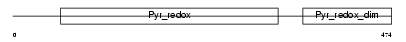
|
|||||
| Description | Glutathione reductase, mitochondrial precursor (EC 1.8.1.7) (GR) (GRase). | |||||
|
MAGAA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.009919 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43363 | Gene names | MAGEA10, MAGE10 | |||
|
Domain Architecture |
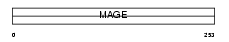
|
|||||
| Description | Melanoma-associated antigen 10 (MAGE-10 antigen). | |||||
|
2ACA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.011610 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06190, Q06189, Q9NPQ5 | Gene names | PPP2R3A, PPP2R3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A 72/130 kDa regulatory subunit B (PP2A, subunit B, B''-PR72/PR130) (PP2A, subunit B, B72/B130 isoforms) (PP2A, subunit B, PR72/PR130 isoforms) (PP2A, subunit B, R3 isoform). | |||||
|
FMO3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.048778 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P97501 | Gene names | Fmo3 | |||
|
Domain Architecture |
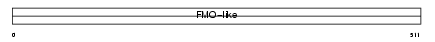
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 3 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 3) (FMO 3) (Dimethylaniline oxidase 3). | |||||
|
N4BP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.010151 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15049, Q7Z6I3 | Gene names | N4BP3, KIAA0341 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 3 (N4BP3). | |||||
|
AMID_MOUSE
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.019049 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BUE4, Q8CHZ2 | Gene names | Amid | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-inducing factor-like mitochondrion-associated inducer of death (EC 1.-.-.-) (Apoptosis-inducing factor-homologous mitochondrion-associated inducer of death). | |||||
|
DHSA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.020541 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31040, Q16395, Q9UMY5 | Gene names | SDHA, SDH2, SDHF | |||
|
Domain Architecture |

|
|||||
| Description | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial precursor (EC 1.3.5.1) (Fp) (Flavoprotein subunit of complex II). | |||||
|
GSTT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.009289 (rank : 38) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61133, Q61134, Q64472 | Gene names | Gstt2 | |||
|
Domain Architecture |
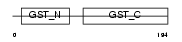
|
|||||
| Description | Glutathione S-transferase theta-2 (EC 2.5.1.18) (GST class-theta-2). | |||||
|
PCYOX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.151999 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CQF9, Q3UHV6, Q69ZW0, Q8BZX1 | Gene names | Pcyox1, Kiaa0908 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prenylcysteine oxidase precursor (EC 1.8.3.5). | |||||
|
AOFA_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q64133, Q8K0Z8 | Gene names | Maoa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A). | |||||
|
AOFB_HUMAN
|
||||||
| NC score | 0.989564 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P27338, Q7Z6S2 | Gene names | MAOB | |||
|
Domain Architecture |
|
|||||
| Description | Amine oxidase [flavin-containing] B (EC 1.4.3.4) (Monoamine oxidase type B) (MAO-B). | |||||
|
AOFB_MOUSE
|
||||||
| NC score | 0.987861 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BW75, Q8C0B2 | Gene names | Maob | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amine oxidase [flavin-containing] B (EC 1.4.3.4) (Monoamine oxidase type B) (MAO-B). | |||||
|
AOFA_HUMAN
|
||||||
| NC score | 0.987545 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P21397, Q16426 | Gene names | MAOA | |||
|
Domain Architecture |

|
|||||
| Description | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A). | |||||
|
AOF1_HUMAN
|
||||||
| NC score | 0.547626 (rank : 5) | θ value | 1.52774e-15 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NB78, Q5TGV3, Q6AI15, Q6ZUU4, Q8N258, Q96EL7 | Gene names | AOF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
AOF1_MOUSE
|
||||||
| NC score | 0.543833 (rank : 6) | θ value | 1.68911e-14 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CIG3, Q8C5C4, Q8CEC1 | Gene names | Aof1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
OXLA_MOUSE
|
||||||
| NC score | 0.499876 (rank : 7) | θ value | 4.30538e-10 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O09046, Q3T9N9, Q3U7S6, Q3V0K2, Q6Y632, Q6YBV6, Q6YDI8, Q8R2G8, Q9CXK7 | Gene names | Il4i1, Fig1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (mFIG1). | |||||
|
OXLA_HUMAN
|
||||||
| NC score | 0.470552 (rank : 8) | θ value | 7.34386e-10 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96RQ9, Q4GZN1, Q4GZN2, Q6P2Q3, Q8TEM5, Q96RQ8 | Gene names | IL4I1, FIG1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (hFIG1). | |||||
|
LSD1_MOUSE
|
||||||
| NC score | 0.420796 (rank : 9) | θ value | 2.79066e-09 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZQ88, Q6PB53, Q8VEA1 | Gene names | Aof2, Kiaa0601, Lsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
PAOX_MOUSE
|
||||||
| NC score | 0.415194 (rank : 10) | θ value | 0.00035302 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C0L6, Q8K254 | Gene names | Paox, Pao | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal N1-acetyl-spermine/spermidine oxidase (EC 1.5.3.11) (Polyamine oxidase). | |||||
|
LSD1_HUMAN
|
||||||
| NC score | 0.403246 (rank : 11) | θ value | 9.59137e-10 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
PAOX_HUMAN
|
||||||
| NC score | 0.333806 (rank : 12) | θ value | 0.0148317 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6QHF9, Q6QHF5, Q6QHF6, Q6QHF7, Q6QHF8, Q6QHG0, Q6QHG1, Q6QHG2, Q6QHG3, Q6QHG4, Q6QHG5, Q6QHG6, Q86WP9, Q8N555, Q8NCX3 | Gene names | PAOX, PAO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal N1-acetyl-spermine/spermidine oxidase (EC 1.5.3.11) (Polyamine oxidase). | |||||
|
SMOX_HUMAN
|
||||||
| NC score | 0.330025 (rank : 13) | θ value | 0.00020696 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NWM0, Q5TE26, Q5TE27, Q6UY28, Q8IX00, Q96LC3, Q96LC4, Q96QT3, Q9BW38, Q9H6H1, Q9NP51, Q9NPY1, Q9NPY2 | Gene names | SMOX, C20orf16, SMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermine oxidase (EC 1.5.3.-) (Polyamine oxidase 1) (PAO-1) (PAOh1). | |||||
|
SMOX_MOUSE
|
||||||
| NC score | 0.320009 (rank : 14) | θ value | 0.00175202 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99K82, Q70LA3, Q70LA4, Q70LA5, Q70LA7, Q70LA8, Q70LA9, Q70LB0, Q8CJ56, Q8CJ57 | Gene names | Smox, Smo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermine oxidase (EC 1.5.3.-) (Polyamine oxidase 1) (PAO-1) (PAOh1). | |||||
|
PPOX_HUMAN
|
||||||
| NC score | 0.259996 (rank : 15) | θ value | 0.00390308 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P50336 | Gene names | PPOX | |||
|
Domain Architecture |
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
CJ033_MOUSE
|
||||||
| NC score | 0.184408 (rank : 16) | θ value | 0.0736092 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3U4I7, Q8CAN2, Q9D630 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 homolog (EC 1.-.-.-). | |||||
|
RETST_HUMAN
|
||||||
| NC score | 0.182123 (rank : 17) | θ value | 0.000786445 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6NUM9, Q53R95, Q53SA9, Q6UX05, Q8N2H5, Q96FA4, Q9NXE5 | Gene names | RETSAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
PPOX_MOUSE
|
||||||
| NC score | 0.180940 (rank : 18) | θ value | 0.0330416 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P51175, P97344, Q99M34 | Gene names | Ppox | |||
|
Domain Architecture |
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
CJ033_HUMAN
|
||||||
| NC score | 0.175553 (rank : 19) | θ value | 0.0736092 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N2H3, Q5TAA9, Q9BRQ1 | Gene names | C10orf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 (EC 1.-.-.-). | |||||
|
RETST_MOUSE
|
||||||
| NC score | 0.173162 (rank : 20) | θ value | 0.00228821 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64FW2, Q3UNN9, Q78JX8, Q8VHE7, Q9CW85 | Gene names | Retsat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
PCYOX_MOUSE
|
||||||
| NC score | 0.151999 (rank : 21) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CQF9, Q3UHV6, Q69ZW0, Q8BZX1 | Gene names | Pcyox1, Kiaa0908 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prenylcysteine oxidase precursor (EC 1.8.3.5). | |||||
|
PCYOX_HUMAN
|
||||||
| NC score | 0.105419 (rank : 22) | θ value | 0.0330416 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHG3, O94982, Q8N4N5, Q96QM8 | Gene names | PCYOX1, KIAA0908, PCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Prenylcysteine oxidase precursor (EC 1.8.3.5) (PCL1). | |||||
|
TRXR1_HUMAN
|
||||||
| NC score | 0.058439 (rank : 23) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16881, Q76P53, Q7LA96, Q9UH79 | Gene names | TXNRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 1, cytoplasmic precursor (EC 1.8.1.9) (TR) (TR1). | |||||
|
TRXR1_MOUSE
|
||||||
| NC score | 0.053479 (rank : 24) | θ value | 0.125558 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JMH6 | Gene names | Txnrd1, Trxr1 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 1, cytoplasmic (EC 1.8.1.9) (TR) (TR1). | |||||
|
FMO3_MOUSE
|
||||||
| NC score | 0.048778 (rank : 25) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P97501 | Gene names | Fmo3 | |||
|
Domain Architecture |
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 3 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 3) (FMO 3) (Dimethylaniline oxidase 3). | |||||
|
GSHR_MOUSE
|
||||||
| NC score | 0.047266 (rank : 26) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P47791, Q7TNC2, Q8BN97, Q8C9Z6 | Gene names | Gsr, Gr1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione reductase, mitochondrial precursor (EC 1.8.1.7) (GR) (GRase). | |||||
|
L2HDH_MOUSE
|
||||||
| NC score | 0.043678 (rank : 27) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91YP0, Q3TH61, Q3U7Z0, Q3ULY6 | Gene names | L2hgdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L-2-hydroxyglutarate dehydrogenase, mitochondrial precursor (EC 1.1.99.2) (Duranin). | |||||
|
GSHR_HUMAN
|
||||||
| NC score | 0.043143 (rank : 28) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P00390, Q7Z5C9, Q9NP63 | Gene names | GSR, GLUR, GRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione reductase, mitochondrial precursor (EC 1.8.1.7) (GR) (GRase). | |||||
|
L2HDH_HUMAN
|
||||||
| NC score | 0.042824 (rank : 29) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H9P8, Q9BRR1 | Gene names | L2HGDH, C14orf160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L-2-hydroxyglutarate dehydrogenase, mitochondrial precursor (EC 1.1.99.2) (Duranin). | |||||
|
FMO5_HUMAN
|
||||||
| NC score | 0.024604 (rank : 30) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49326 | Gene names | FMO5 | |||
|
Domain Architecture |
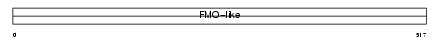
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 5 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 5) (FMO 5) (Dimethylaniline oxidase 5). | |||||
|
DHSA_MOUSE
|
||||||
| NC score | 0.022939 (rank : 31) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K2B3, Q3UH25 | Gene names | Sdha | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial precursor (EC 1.3.5.1) (Fp) (Flavoprotein subunit of complex II). | |||||
|
AMID_HUMAN
|
||||||
| NC score | 0.021771 (rank : 32) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BRQ8, Q63Z39 | Gene names | AMID, PRG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-inducing factor-like mitochondrion-associated inducer of death (EC 1.-.-.-) (Apoptosis-inducing factor-homologous mitochondrion-associated inducer of death) (p53-responsive gene 3 protein). | |||||
|
DHSA_HUMAN
|
||||||
| NC score | 0.020541 (rank : 33) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31040, Q16395, Q9UMY5 | Gene names | SDHA, SDH2, SDHF | |||
|
Domain Architecture |

|
|||||
| Description | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial precursor (EC 1.3.5.1) (Fp) (Flavoprotein subunit of complex II). | |||||
|
AMID_MOUSE
|
||||||
| NC score | 0.019049 (rank : 34) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BUE4, Q8CHZ2 | Gene names | Amid | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-inducing factor-like mitochondrion-associated inducer of death (EC 1.-.-.-) (Apoptosis-inducing factor-homologous mitochondrion-associated inducer of death). | |||||
|
2ACA_HUMAN
|
||||||
| NC score | 0.011610 (rank : 35) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06190, Q06189, Q9NPQ5 | Gene names | PPP2R3A, PPP2R3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A 72/130 kDa regulatory subunit B (PP2A, subunit B, B''-PR72/PR130) (PP2A, subunit B, B72/B130 isoforms) (PP2A, subunit B, PR72/PR130 isoforms) (PP2A, subunit B, R3 isoform). | |||||
|
N4BP3_HUMAN
|
||||||
| NC score | 0.010151 (rank : 36) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15049, Q7Z6I3 | Gene names | N4BP3, KIAA0341 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 3 (N4BP3). | |||||
|
MAGAA_HUMAN
|
||||||
| NC score | 0.009919 (rank : 37) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43363 | Gene names | MAGEA10, MAGE10 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 10 (MAGE-10 antigen). | |||||
|
GSTT2_MOUSE
|
||||||
| NC score | 0.009289 (rank : 38) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61133, Q61134, Q64472 | Gene names | Gstt2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione S-transferase theta-2 (EC 2.5.1.18) (GST class-theta-2). | |||||