Please be patient as the page loads
|
DLDH_HUMAN
|
||||||
| SwissProt Accessions | P09622, Q14131, Q14167 | Gene names | DLD, GCSL, LAD, PHE3 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase) (Glycine cleavage system L protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DLDH_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P09622, Q14131, Q14167 | Gene names | DLD, GCSL, LAD, PHE3 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase) (Glycine cleavage system L protein). | |||||
|
DLDH_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998979 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O08749 | Gene names | Dld | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase). | |||||
|
TRXR1_MOUSE
|
||||||
| θ value | 1.36902e-48 (rank : 3) | NC score | 0.874251 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JMH6 | Gene names | Txnrd1, Trxr1 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 1, cytoplasmic (EC 1.8.1.9) (TR) (TR1). | |||||
|
TRXR1_HUMAN
|
||||||
| θ value | 2.3352e-48 (rank : 4) | NC score | 0.872623 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16881, Q76P53, Q7LA96, Q9UH79 | Gene names | TXNRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 1, cytoplasmic precursor (EC 1.8.1.9) (TR) (TR1). | |||||
|
GSHR_HUMAN
|
||||||
| θ value | 3.15612e-45 (rank : 5) | NC score | 0.877917 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00390, Q7Z5C9, Q9NP63 | Gene names | GSR, GLUR, GRD1 | |||
|
Domain Architecture |
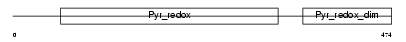
|
|||||
| Description | Glutathione reductase, mitochondrial precursor (EC 1.8.1.7) (GR) (GRase). | |||||
|
TRXR2_HUMAN
|
||||||
| θ value | 9.18288e-45 (rank : 6) | NC score | 0.868126 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NNW7, O95840, Q96IJ2, Q9H2Z5, Q9NZV3, Q9NZV4, Q9P2Y0, Q9P2Y1, Q9UQU8 | Gene names | TXNRD2, TRXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 2, mitochondrial precursor (EC 1.8.1.9) (TR3) (TR-beta) (Selenoprotein Z) (SelZ). | |||||
|
GSHR_MOUSE
|
||||||
| θ value | 8.59492e-43 (rank : 7) | NC score | 0.871161 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P47791, Q7TNC2, Q8BN97, Q8C9Z6 | Gene names | Gsr, Gr1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione reductase, mitochondrial precursor (EC 1.8.1.7) (GR) (GRase). | |||||
|
TRXR2_MOUSE
|
||||||
| θ value | 7.27602e-42 (rank : 8) | NC score | 0.864803 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JLT4, Q91YX4, Q9JHA7, Q9JMH5 | Gene names | Txnrd2, Trxr2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 2, mitochondrial precursor (EC 1.8.1.9) (TR3). | |||||
|
RETST_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 9) | NC score | 0.118015 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6NUM9, Q53R95, Q53SA9, Q6UX05, Q8N2H5, Q96FA4, Q9NXE5 | Gene names | RETSAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
CJ033_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.136069 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N2H3, Q5TAA9, Q9BRQ1 | Gene names | C10orf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 (EC 1.-.-.-). | |||||
|
CJ033_MOUSE
|
||||||
| θ value | 0.21417 (rank : 11) | NC score | 0.128424 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3U4I7, Q8CAN2, Q9D630 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 homolog (EC 1.-.-.-). | |||||
|
RETST_MOUSE
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.107930 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q64FW2, Q3UNN9, Q78JX8, Q8VHE7, Q9CW85 | Gene names | Retsat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
ETFD_MOUSE
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.082554 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q921G7, Q3U7K2, Q8BK82, Q9DCT9 | Gene names | Etfdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial precursor (EC 1.5.5.1) (ETF-QO) (ETF-ubiquinone oxidoreductase) (ETF dehydrogenase) (Electron-transferring- flavoprotein dehydrogenase). | |||||
|
ETFD_HUMAN
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.082785 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16134, Q7Z347 | Gene names | ETFDH | |||
|
Domain Architecture |

|
|||||
| Description | Electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial precursor (EC 1.5.5.1) (ETF-QO) (ETF-ubiquinone oxidoreductase) (ETF dehydrogenase) (Electron-transferring- flavoprotein dehydrogenase). | |||||
|
LSD1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.054208 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6ZQ88, Q6PB53, Q8VEA1 | Gene names | Aof2, Kiaa0601, Lsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
OXLA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.068093 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O09046, Q3T9N9, Q3U7S6, Q3V0K2, Q6Y632, Q6YBV6, Q6YDI8, Q8R2G8, Q9CXK7 | Gene names | Il4i1, Fig1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (mFIG1). | |||||
|
ERG1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 17) | NC score | 0.058322 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P52019 | Gene names | Sqle, Erg1 | |||
|
Domain Architecture |
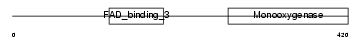
|
|||||
| Description | Squalene monooxygenase (EC 1.14.99.7) (Squalene epoxidase) (SE). | |||||
|
LSD1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.049893 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
MICA2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.030567 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BML1, Q3UPU6 | Gene names | Mical2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
OXLA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.076707 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96RQ9, Q4GZN1, Q4GZN2, Q6P2Q3, Q8TEM5, Q96RQ8 | Gene names | IL4I1, FIG1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (hFIG1). | |||||
|
ERG1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.054939 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14534, Q9UEK6 | Gene names | SQLE, ERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Squalene monooxygenase (EC 1.14.99.7) (Squalene epoxidase) (SE). | |||||
|
AOF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.052858 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NB78, Q5TGV3, Q6AI15, Q6ZUU4, Q8N258, Q96EL7 | Gene names | AOF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
AOF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.051698 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CIG3, Q8C5C4, Q8CEC1 | Gene names | Aof1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
CO5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.016750 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01031 | Gene names | C5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C5 precursor [Contains: Complement C5 beta chain; Complement C5 alpha chain; C5a anaphylatoxin; Complement C5 alpha' chain]. | |||||
|
GPDM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.071258 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64521, Q61507 | Gene names | Gpd2, Gdm1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M). | |||||
|
DHSA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.039332 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31040, Q16395, Q9UMY5 | Gene names | SDHA, SDH2, SDHF | |||
|
Domain Architecture |

|
|||||
| Description | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial precursor (EC 1.3.5.1) (Fp) (Flavoprotein subunit of complex II). | |||||
|
GPDM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.068525 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P43304, Q59FR1, Q9HAP9 | Gene names | GPD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M) (mtGPD). | |||||
|
MICA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.024962 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94851, Q7Z3A8 | Gene names | MICAL2, KIAA0750 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
CO5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.019562 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06684 | Gene names | C5, Hc | |||
|
Domain Architecture |

|
|||||
| Description | Complement C5 precursor (Hemolytic complement) [Contains: Complement C5 beta chain; Complement C5 alpha chain; C5a anaphylatoxin; Complement C5 alpha' chain]. | |||||
|
DHSA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.032222 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K2B3, Q3UH25 | Gene names | Sdha | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial precursor (EC 1.3.5.1) (Fp) (Flavoprotein subunit of complex II). | |||||
|
PYR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.014262 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
SARDH_MOUSE
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.026859 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99LB7 | Gene names | Sardh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcosine dehydrogenase, mitochondrial precursor (EC 1.5.99.1) (SarDH). | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.002886 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
MICA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.014884 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VDP3 | Gene names | Mical1, Mical, Nical | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
DLDH_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P09622, Q14131, Q14167 | Gene names | DLD, GCSL, LAD, PHE3 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase) (Glycine cleavage system L protein). | |||||
|
DLDH_MOUSE
|
||||||
| NC score | 0.998979 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O08749 | Gene names | Dld | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase). | |||||
|
GSHR_HUMAN
|
||||||
| NC score | 0.877917 (rank : 3) | θ value | 3.15612e-45 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00390, Q7Z5C9, Q9NP63 | Gene names | GSR, GLUR, GRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione reductase, mitochondrial precursor (EC 1.8.1.7) (GR) (GRase). | |||||
|
TRXR1_MOUSE
|
||||||
| NC score | 0.874251 (rank : 4) | θ value | 1.36902e-48 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JMH6 | Gene names | Txnrd1, Trxr1 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 1, cytoplasmic (EC 1.8.1.9) (TR) (TR1). | |||||
|
TRXR1_HUMAN
|
||||||
| NC score | 0.872623 (rank : 5) | θ value | 2.3352e-48 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16881, Q76P53, Q7LA96, Q9UH79 | Gene names | TXNRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 1, cytoplasmic precursor (EC 1.8.1.9) (TR) (TR1). | |||||
|
GSHR_MOUSE
|
||||||
| NC score | 0.871161 (rank : 6) | θ value | 8.59492e-43 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P47791, Q7TNC2, Q8BN97, Q8C9Z6 | Gene names | Gsr, Gr1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione reductase, mitochondrial precursor (EC 1.8.1.7) (GR) (GRase). | |||||
|
TRXR2_HUMAN
|
||||||
| NC score | 0.868126 (rank : 7) | θ value | 9.18288e-45 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NNW7, O95840, Q96IJ2, Q9H2Z5, Q9NZV3, Q9NZV4, Q9P2Y0, Q9P2Y1, Q9UQU8 | Gene names | TXNRD2, TRXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 2, mitochondrial precursor (EC 1.8.1.9) (TR3) (TR-beta) (Selenoprotein Z) (SelZ). | |||||
|
TRXR2_MOUSE
|
||||||
| NC score | 0.864803 (rank : 8) | θ value | 7.27602e-42 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JLT4, Q91YX4, Q9JHA7, Q9JMH5 | Gene names | Txnrd2, Trxr2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 2, mitochondrial precursor (EC 1.8.1.9) (TR3). | |||||
|
CJ033_HUMAN
|
||||||
| NC score | 0.136069 (rank : 9) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N2H3, Q5TAA9, Q9BRQ1 | Gene names | C10orf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 (EC 1.-.-.-). | |||||
|
CJ033_MOUSE
|
||||||
| NC score | 0.128424 (rank : 10) | θ value | 0.21417 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3U4I7, Q8CAN2, Q9D630 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 homolog (EC 1.-.-.-). | |||||
|
RETST_HUMAN
|
||||||
| NC score | 0.118015 (rank : 11) | θ value | 0.0736092 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6NUM9, Q53R95, Q53SA9, Q6UX05, Q8N2H5, Q96FA4, Q9NXE5 | Gene names | RETSAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
RETST_MOUSE
|
||||||
| NC score | 0.107930 (rank : 12) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q64FW2, Q3UNN9, Q78JX8, Q8VHE7, Q9CW85 | Gene names | Retsat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | All-trans-retinol 13,14-reductase precursor (EC 1.3.99.23) (All-trans- 13,14-dihydroretinol saturase) (RetSat). | |||||
|
ETFD_HUMAN
|
||||||
| NC score | 0.082785 (rank : 13) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16134, Q7Z347 | Gene names | ETFDH | |||
|
Domain Architecture |

|
|||||
| Description | Electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial precursor (EC 1.5.5.1) (ETF-QO) (ETF-ubiquinone oxidoreductase) (ETF dehydrogenase) (Electron-transferring- flavoprotein dehydrogenase). | |||||
|
ETFD_MOUSE
|
||||||
| NC score | 0.082554 (rank : 14) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q921G7, Q3U7K2, Q8BK82, Q9DCT9 | Gene names | Etfdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial precursor (EC 1.5.5.1) (ETF-QO) (ETF-ubiquinone oxidoreductase) (ETF dehydrogenase) (Electron-transferring- flavoprotein dehydrogenase). | |||||
|
OXLA_HUMAN
|
||||||
| NC score | 0.076707 (rank : 15) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96RQ9, Q4GZN1, Q4GZN2, Q6P2Q3, Q8TEM5, Q96RQ8 | Gene names | IL4I1, FIG1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (hFIG1). | |||||
|
GPDM_MOUSE
|
||||||
| NC score | 0.071258 (rank : 16) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64521, Q61507 | Gene names | Gpd2, Gdm1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M). | |||||
|
GPDM_HUMAN
|
||||||
| NC score | 0.068525 (rank : 17) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P43304, Q59FR1, Q9HAP9 | Gene names | GPD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M) (mtGPD). | |||||
|
OXLA_MOUSE
|
||||||
| NC score | 0.068093 (rank : 18) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O09046, Q3T9N9, Q3U7S6, Q3V0K2, Q6Y632, Q6YBV6, Q6YDI8, Q8R2G8, Q9CXK7 | Gene names | Il4i1, Fig1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (mFIG1). | |||||
|
ERG1_MOUSE
|
||||||
| NC score | 0.058322 (rank : 19) | θ value | 1.81305 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P52019 | Gene names | Sqle, Erg1 | |||
|
Domain Architecture |

|
|||||
| Description | Squalene monooxygenase (EC 1.14.99.7) (Squalene epoxidase) (SE). | |||||
|
ERG1_HUMAN
|
||||||
| NC score | 0.054939 (rank : 20) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14534, Q9UEK6 | Gene names | SQLE, ERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Squalene monooxygenase (EC 1.14.99.7) (Squalene epoxidase) (SE). | |||||
|
LSD1_MOUSE
|
||||||
| NC score | 0.054208 (rank : 21) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6ZQ88, Q6PB53, Q8VEA1 | Gene names | Aof2, Kiaa0601, Lsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
AOF1_HUMAN
|
||||||
| NC score | 0.052858 (rank : 22) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NB78, Q5TGV3, Q6AI15, Q6ZUU4, Q8N258, Q96EL7 | Gene names | AOF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
AOF1_MOUSE
|
||||||
| NC score | 0.051698 (rank : 23) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CIG3, Q8C5C4, Q8CEC1 | Gene names | Aof1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
LSD1_HUMAN
|
||||||
| NC score | 0.049893 (rank : 24) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
DHSA_HUMAN
|
||||||
| NC score | 0.039332 (rank : 25) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31040, Q16395, Q9UMY5 | Gene names | SDHA, SDH2, SDHF | |||
|
Domain Architecture |

|
|||||
| Description | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial precursor (EC 1.3.5.1) (Fp) (Flavoprotein subunit of complex II). | |||||
|
DHSA_MOUSE
|
||||||
| NC score | 0.032222 (rank : 26) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K2B3, Q3UH25 | Gene names | Sdha | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial precursor (EC 1.3.5.1) (Fp) (Flavoprotein subunit of complex II). | |||||
|
MICA2_MOUSE
|
||||||
| NC score | 0.030567 (rank : 27) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BML1, Q3UPU6 | Gene names | Mical2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
SARDH_MOUSE
|
||||||
| NC score | 0.026859 (rank : 28) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99LB7 | Gene names | Sardh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcosine dehydrogenase, mitochondrial precursor (EC 1.5.99.1) (SarDH). | |||||
|
MICA2_HUMAN
|
||||||
| NC score | 0.024962 (rank : 29) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94851, Q7Z3A8 | Gene names | MICAL2, KIAA0750 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
CO5_MOUSE
|
||||||
| NC score | 0.019562 (rank : 30) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06684 | Gene names | C5, Hc | |||
|
Domain Architecture |

|
|||||
| Description | Complement C5 precursor (Hemolytic complement) [Contains: Complement C5 beta chain; Complement C5 alpha chain; C5a anaphylatoxin; Complement C5 alpha' chain]. | |||||
|
CO5_HUMAN
|
||||||
| NC score | 0.016750 (rank : 31) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01031 | Gene names | C5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C5 precursor [Contains: Complement C5 beta chain; Complement C5 alpha chain; C5a anaphylatoxin; Complement C5 alpha' chain]. | |||||
|
MICA1_MOUSE
|
||||||
| NC score | 0.014884 (rank : 32) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VDP3 | Gene names | Mical1, Mical, Nical | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
PYR1_HUMAN
|
||||||
| NC score | 0.014262 (rank : 33) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.002886 (rank : 34) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||