Please be patient as the page loads
|
TRXR2_HUMAN
|
||||||
| SwissProt Accessions | Q9NNW7, O95840, Q96IJ2, Q9H2Z5, Q9NZV3, Q9NZV4, Q9P2Y0, Q9P2Y1, Q9UQU8 | Gene names | TXNRD2, TRXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 2, mitochondrial precursor (EC 1.8.1.9) (TR3) (TR-beta) (Selenoprotein Z) (SelZ). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TRXR2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NNW7, O95840, Q96IJ2, Q9H2Z5, Q9NZV3, Q9NZV4, Q9P2Y0, Q9P2Y1, Q9UQU8 | Gene names | TXNRD2, TRXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 2, mitochondrial precursor (EC 1.8.1.9) (TR3) (TR-beta) (Selenoprotein Z) (SelZ). | |||||
|
TRXR2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998186 (rank : 2) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JLT4, Q91YX4, Q9JHA7, Q9JMH5 | Gene names | Txnrd2, Trxr2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 2, mitochondrial precursor (EC 1.8.1.9) (TR3). | |||||
|
TRXR1_MOUSE
|
||||||
| θ value | 3.73483e-155 (rank : 3) | NC score | 0.987040 (rank : 3) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JMH6 | Gene names | Txnrd1, Trxr1 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 1, cytoplasmic (EC 1.8.1.9) (TR) (TR1). | |||||
|
TRXR1_HUMAN
|
||||||
| θ value | 5.96274e-153 (rank : 4) | NC score | 0.986632 (rank : 4) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q16881, Q76P53, Q7LA96, Q9UH79 | Gene names | TXNRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 1, cytoplasmic precursor (EC 1.8.1.9) (TR) (TR1). | |||||
|
GSHR_HUMAN
|
||||||
| θ value | 3.82246e-75 (rank : 5) | NC score | 0.939858 (rank : 5) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P00390, Q7Z5C9, Q9NP63 | Gene names | GSR, GLUR, GRD1 | |||
|
Domain Architecture |
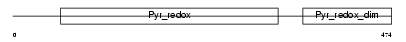
|
|||||
| Description | Glutathione reductase, mitochondrial precursor (EC 1.8.1.7) (GR) (GRase). | |||||
|
GSHR_MOUSE
|
||||||
| θ value | 8.51556e-75 (rank : 6) | NC score | 0.939599 (rank : 6) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P47791, Q7TNC2, Q8BN97, Q8C9Z6 | Gene names | Gsr, Gr1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione reductase, mitochondrial precursor (EC 1.8.1.7) (GR) (GRase). | |||||
|
DLDH_HUMAN
|
||||||
| θ value | 9.18288e-45 (rank : 7) | NC score | 0.868126 (rank : 7) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P09622, Q14131, Q14167 | Gene names | DLD, GCSL, LAD, PHE3 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase) (Glycine cleavage system L protein). | |||||
|
DLDH_MOUSE
|
||||||
| θ value | 3.48949e-44 (rank : 8) | NC score | 0.866169 (rank : 8) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O08749 | Gene names | Dld | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase). | |||||
|
ADPN_HUMAN
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.085834 (rank : 11) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NST1, Q6P1A1, Q96CB4 | Gene names | PNPLA3, ADPN, C22orf20 | |||
|
Domain Architecture |
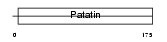
|
|||||
| Description | Adiponutrin (iPLA2-epsilon) (Calcium-independent phospholipase A2- epsilon) (Patatin-like phospholipase domain-containing protein 3) [Includes: Triacylglycerol lipase (EC 3.1.1.3); Acylglycerol O- acyltransferase (EC 2.3.1.-)]. | |||||
|
GPDM_MOUSE
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.093653 (rank : 10) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64521, Q61507 | Gene names | Gpd2, Gdm1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M). | |||||
|
GPDM_HUMAN
|
||||||
| θ value | 0.21417 (rank : 11) | NC score | 0.097365 (rank : 9) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P43304, Q59FR1, Q9HAP9 | Gene names | GPD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M) (mtGPD). | |||||
|
SQRD_HUMAN
|
||||||
| θ value | 0.62314 (rank : 12) | NC score | 0.050044 (rank : 14) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6N5, Q9UQM8 | Gene names | SQRDL | |||
|
Domain Architecture |
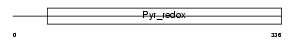
|
|||||
| Description | Sulfide:quinone oxidoreductase, mitochondrial precursor (EC 1.-.-.-). | |||||
|
IMA2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.012717 (rank : 17) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P52292, Q9BRU5 | Gene names | KPNA2, RCH1, SRP1 | |||
|
Domain Architecture |
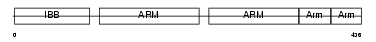
|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1). | |||||
|
MICA2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.022994 (rank : 15) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BML1, Q3UPU6 | Gene names | Mical2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
MICA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.019964 (rank : 16) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O94851, Q7Z3A8 | Gene names | MICAL2, KIAA0750 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
CJ033_HUMAN
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.068786 (rank : 12) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N2H3, Q5TAA9, Q9BRQ1 | Gene names | C10orf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 (EC 1.-.-.-). | |||||
|
CJ033_MOUSE
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.061034 (rank : 13) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3U4I7, Q8CAN2, Q9D630 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 homolog (EC 1.-.-.-). | |||||
|
CHSS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 18) | NC score | 0.005918 (rank : 18) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
TRXR2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NNW7, O95840, Q96IJ2, Q9H2Z5, Q9NZV3, Q9NZV4, Q9P2Y0, Q9P2Y1, Q9UQU8 | Gene names | TXNRD2, TRXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 2, mitochondrial precursor (EC 1.8.1.9) (TR3) (TR-beta) (Selenoprotein Z) (SelZ). | |||||
|
TRXR2_MOUSE
|
||||||
| NC score | 0.998186 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JLT4, Q91YX4, Q9JHA7, Q9JMH5 | Gene names | Txnrd2, Trxr2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 2, mitochondrial precursor (EC 1.8.1.9) (TR3). | |||||
|
TRXR1_MOUSE
|
||||||
| NC score | 0.987040 (rank : 3) | θ value | 3.73483e-155 (rank : 3) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JMH6 | Gene names | Txnrd1, Trxr1 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 1, cytoplasmic (EC 1.8.1.9) (TR) (TR1). | |||||
|
TRXR1_HUMAN
|
||||||
| NC score | 0.986632 (rank : 4) | θ value | 5.96274e-153 (rank : 4) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q16881, Q76P53, Q7LA96, Q9UH79 | Gene names | TXNRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 1, cytoplasmic precursor (EC 1.8.1.9) (TR) (TR1). | |||||
|
GSHR_HUMAN
|
||||||
| NC score | 0.939858 (rank : 5) | θ value | 3.82246e-75 (rank : 5) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P00390, Q7Z5C9, Q9NP63 | Gene names | GSR, GLUR, GRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione reductase, mitochondrial precursor (EC 1.8.1.7) (GR) (GRase). | |||||
|
GSHR_MOUSE
|
||||||
| NC score | 0.939599 (rank : 6) | θ value | 8.51556e-75 (rank : 6) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P47791, Q7TNC2, Q8BN97, Q8C9Z6 | Gene names | Gsr, Gr1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione reductase, mitochondrial precursor (EC 1.8.1.7) (GR) (GRase). | |||||
|
DLDH_HUMAN
|
||||||
| NC score | 0.868126 (rank : 7) | θ value | 9.18288e-45 (rank : 7) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P09622, Q14131, Q14167 | Gene names | DLD, GCSL, LAD, PHE3 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase) (Glycine cleavage system L protein). | |||||
|
DLDH_MOUSE
|
||||||
| NC score | 0.866169 (rank : 8) | θ value | 3.48949e-44 (rank : 8) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O08749 | Gene names | Dld | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase). | |||||
|
GPDM_HUMAN
|
||||||
| NC score | 0.097365 (rank : 9) | θ value | 0.21417 (rank : 11) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P43304, Q59FR1, Q9HAP9 | Gene names | GPD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M) (mtGPD). | |||||
|
GPDM_MOUSE
|
||||||
| NC score | 0.093653 (rank : 10) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64521, Q61507 | Gene names | Gpd2, Gdm1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M). | |||||
|
ADPN_HUMAN
|
||||||
| NC score | 0.085834 (rank : 11) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NST1, Q6P1A1, Q96CB4 | Gene names | PNPLA3, ADPN, C22orf20 | |||
|
Domain Architecture |

|
|||||
| Description | Adiponutrin (iPLA2-epsilon) (Calcium-independent phospholipase A2- epsilon) (Patatin-like phospholipase domain-containing protein 3) [Includes: Triacylglycerol lipase (EC 3.1.1.3); Acylglycerol O- acyltransferase (EC 2.3.1.-)]. | |||||
|
CJ033_HUMAN
|
||||||
| NC score | 0.068786 (rank : 12) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N2H3, Q5TAA9, Q9BRQ1 | Gene names | C10orf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 (EC 1.-.-.-). | |||||
|
CJ033_MOUSE
|
||||||
| NC score | 0.061034 (rank : 13) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3U4I7, Q8CAN2, Q9D630 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 homolog (EC 1.-.-.-). | |||||
|
SQRD_HUMAN
|
||||||
| NC score | 0.050044 (rank : 14) | θ value | 0.62314 (rank : 12) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6N5, Q9UQM8 | Gene names | SQRDL | |||
|
Domain Architecture |

|
|||||
| Description | Sulfide:quinone oxidoreductase, mitochondrial precursor (EC 1.-.-.-). | |||||
|
MICA2_MOUSE
|
||||||
| NC score | 0.022994 (rank : 15) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BML1, Q3UPU6 | Gene names | Mical2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
MICA2_HUMAN
|
||||||
| NC score | 0.019964 (rank : 16) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O94851, Q7Z3A8 | Gene names | MICAL2, KIAA0750 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
IMA2_HUMAN
|
||||||
| NC score | 0.012717 (rank : 17) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P52292, Q9BRU5 | Gene names | KPNA2, RCH1, SRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1). | |||||
|
CHSS1_MOUSE
|
||||||
| NC score | 0.005918 (rank : 18) | θ value | 8.99809 (rank : 18) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||