Please be patient as the page loads
|
CHSS1_MOUSE
|
||||||
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CHSS1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999192 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q86X52, Q6UX38, Q7LFU5, Q9Y2J5 | Gene names | CHSY1, CHSY, CSS1, KIAA0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase 1) (Chondroitin sulfate synthase 1) (N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS3_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.984780 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
CGAT1_HUMAN
|
||||||
| θ value | 2.3352e-48 (rank : 4) | NC score | 0.805999 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TDX6, Q6P9G6, Q8IUF9, Q9NSQ7, Q9NUM9 | Gene names | GALNACT1, CHGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT1_MOUSE
|
||||||
| θ value | 1.15895e-47 (rank : 5) | NC score | 0.807741 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BJQ9, Q3UZX6, Q8BWV9, Q8BZU7, Q8C195, Q8R0M6 | Gene names | Galnact1, Chgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT2_HUMAN
|
||||||
| θ value | 9.18288e-45 (rank : 6) | NC score | 0.804713 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N6G5, Q6MZJ5, Q6MZP6, Q8TCH4, Q9P1I6 | Gene names | GALNACT2, CHGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CGAT2_MOUSE
|
||||||
| θ value | 2.67181e-44 (rank : 7) | NC score | 0.804179 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8C1F4, Q6PAP6, Q8R5A2, Q9D2M1 | Gene names | Galnact2, Chgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CHSS2_MOUSE
|
||||||
| θ value | 3.05695e-40 (rank : 8) | NC score | 0.716271 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6IQX7 | Gene names | Css2, D1Bwg1363e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase). | |||||
|
CHSS2_HUMAN
|
||||||
| θ value | 3.4976e-36 (rank : 9) | NC score | 0.711708 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZ52, Q6UXD6, Q7L4G1, Q9H0F8, Q9H618 | Gene names | CSS2, CHPF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase) (Chondroitin-polymerizing factor). | |||||
|
CHGUT_HUMAN
|
||||||
| θ value | 1.98606e-31 (rank : 10) | NC score | 0.710797 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2E5, Q6P2I4, Q6UXD2 | Gene names | CSGLCAT, KIAA1402 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate glucuronyltransferase (EC 2.4.1.226) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (Chondroitin glucuronyltransferase II) (CSGlcA-T). | |||||
|
B3GLT_MOUSE
|
||||||
| θ value | 5.43371e-13 (rank : 11) | NC score | 0.401227 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHT6, Q0PCR7, Q149S5 | Gene names | B3galtl, Gm1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
B4GN3_HUMAN
|
||||||
| θ value | 1.58096e-12 (rank : 12) | NC score | 0.477405 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6L9W6, Q6ZNC1, Q8N7T6 | Gene names | B4GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
B4GN3_MOUSE
|
||||||
| θ value | 2.0648e-12 (rank : 13) | NC score | 0.483064 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6L8S8 | Gene names | B4galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
B4GN4_HUMAN
|
||||||
| θ value | 2.69671e-12 (rank : 14) | NC score | 0.489091 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GN4_MOUSE
|
||||||
| θ value | 6.00763e-12 (rank : 15) | NC score | 0.488152 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B3GLT_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 16) | NC score | 0.386581 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6Y288, Q5W0H2, Q6NUI3 | Gene names | B3GALTL, B3GTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
LFNG_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 17) | NC score | 0.261832 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O09010, Q3U659, Q8K3F1, Q9DC10 | Gene names | Lfng | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase lunatic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
LFNG_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 18) | NC score | 0.248611 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NES3, O00589, Q96C39, Q9UJW5 | Gene names | LFNG | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase lunatic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
RFNG_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 19) | NC score | 0.244470 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y644, O00588 | Gene names | RFNG | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase radical fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
RFNG_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 20) | NC score | 0.268525 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09009 | Gene names | Rfng | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase radical fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
B4GT2_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 21) | NC score | 0.192731 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60909, O60511, Q5T4X5, Q5T4Y5, Q9BUP6, Q9NSY7 | Gene names | B4GALT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT2_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 22) | NC score | 0.191344 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Z2Y2, Q3TMP2 | Gene names | B4galt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 23) | NC score | 0.162421 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P15535 | Gene names | B4galt1, Ggtb, Ggtb2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 24) | NC score | 0.151515 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
MFNG_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 25) | NC score | 0.198671 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09008 | Gene names | Mfng | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase manic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
B4GT4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.134953 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60513, Q68D68, Q9BSW3, Q9C078 | Gene names | B4GALT4 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B3GL1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.048218 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75752, Q3Y531, Q6IAI5, Q8NFM8, Q9HA06 | Gene names | B3GALNT1, B3GALT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.79) (Beta-3-GalNAc-T1) (Beta-1,3-galactosyltransferase 3) (Beta-1,3- GalTase 3) (Beta3Gal-T3) (b3Gal-T3) (Galactosylgalactosylglucosylceramide beta-D-acetyl- galactosaminyltransferase) (UDP-N- acetylgalactosamine:globotriaosylceramide beta-1,3-N- acetylgalactosaminyltransferase) (Globoside synthase) (Beta-3-Gx-T3). | |||||
|
MFNG_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.178517 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00587, O43730 | Gene names | MFNG | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase manic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
B4GT4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.138717 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JJ04, Q8BR54, Q9QY12 | Gene names | B4galt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.109408 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O43286, Q2M394, Q9UJQ8 | Gene names | B4GALT5 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.109864 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JMK0 | Gene names | B4galt5, Bgt-5 | |||
|
Domain Architecture |
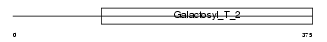
|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.113500 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBX8, O60514 | Gene names | B4GALT6 | |||
|
Domain Architecture |
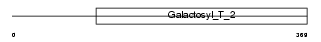
|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.113266 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WVK5 | Gene names | B4galt6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT7_MOUSE
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.111468 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R087, Q7TNU3 | Gene names | B4galt7, Xgalt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
B4GT7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.109019 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UBV7, Q9UHN2 | Gene names | B4GALT7, XGALT1 | |||
|
Domain Architecture |
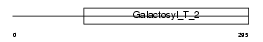
|
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
AHTF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.028214 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CJF7, Q8BVJ5, Q8VD55 | Gene names | Ahctf1, Elys | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
B4GT3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.111615 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60512, O60910, Q9BPZ4, Q9H8T2 | Gene names | B4GALT3 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.112538 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91YY2, Q9QY13 | Gene names | B4galt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
CCDC9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.018564 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
HRB2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.049711 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13601, Q8NEA8, Q8TC37, Q96AT5 | Gene names | HRB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV-1 Rev-binding protein 2 (Rev-interacting protein 1) (Rip-1). | |||||
|
HRB2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.048351 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGA5, Q8C029 | Gene names | Hrb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV-1 Rev-binding protein 2 homolog. | |||||
|
NUDT6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.024088 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53370, O95097, Q9UQD9 | Gene names | NUDT6, FGF2AS | |||
|
Domain Architecture |
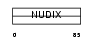
|
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 6 (Nudix motif 6) (Protein GFG) (GFG-1) (Antisense basic fibroblast growth factor). | |||||
|
B3GL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.029297 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q920V1, O54906, Q91V72, Q920V0, Q9CTE5 | Gene names | b3galnt1, B3galt3, B3gt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.79) (Beta-3-GalNAc-T1) (Beta-1,3-galactosyltransferase 3) (Beta-1,3- GalTase 3) (Beta3Gal-T3) (b3Gal-T3) (Galactosylgalactosylglucosylceramide beta-D-acetyl- galactosaminyltransferase) (UDP-N- acetylgalactosamine:globotriaosylceramide beta-1,3-N- acetylgalactosaminyltransferase) (Globoside synthase) (Beta-3-Gx-T3). | |||||
|
FA26B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.023522 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VEC4, Q3TDQ7, Q8BT85, Q8C246 | Gene names | Fam26b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM26B. | |||||
|
LUZP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.003619 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
CCD26_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.023314 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TAB7 | Gene names | CCDC26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 26. | |||||
|
DDAH1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.019975 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CWS0, Q3UF65 | Gene names | Ddah1 | |||
|
Domain Architecture |

|
|||||
| Description | NG,NG-dimethylarginine dimethylaminohydrolase 1 (EC 3.5.3.18) (Dimethylargininase-1) (Dimethylarginine dimethylaminohydrolase 1) (DDAHI) (DDAH-1). | |||||
|
FA26B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.020608 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HA72, O95893, Q6ZUV9 | Gene names | FAM26B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM26B. | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.004643 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
B3GT5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.018491 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2C3, Q9NY96, Q9P1X6, Q9P1X7 | Gene names | B3GALT5 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,3-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,3-GalTase 5) (Beta3Gal-T5) (b3Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,3-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,3- galactosyltransferase 5) (Beta-3-Gx-T5). | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.003598 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
DDAH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.017626 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94760, Q86XK5 | Gene names | DDAH1, DDAH | |||
|
Domain Architecture |

|
|||||
| Description | NG,NG-dimethylarginine dimethylaminohydrolase 1 (EC 3.5.3.18) (Dimethylargininase-1) (Dimethylarginine dimethylaminohydrolase 1) (DDAHI) (DDAH-1). | |||||
|
EMR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.001539 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61549 | Gene names | Emr1, Gpf480 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 1 precursor (Cell surface glycoprotein EMR1) (EMR1 hormone receptor) (Cell surface glycoprotein F4/80). | |||||
|
RBCC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.000194 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
TBX18_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.003047 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPZ6 | Gene names | Tbx18 | |||
|
Domain Architecture |
|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
TRXR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.005918 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NNW7, O95840, Q96IJ2, Q9H2Z5, Q9NZV3, Q9NZV4, Q9P2Y0, Q9P2Y1, Q9UQU8 | Gene names | TXNRD2, TRXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 2, mitochondrial precursor (EC 1.8.1.9) (TR3) (TR-beta) (Selenoprotein Z) (SelZ). | |||||
|
CHSS1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS1_HUMAN
|
||||||
| NC score | 0.999192 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q86X52, Q6UX38, Q7LFU5, Q9Y2J5 | Gene names | CHSY1, CHSY, CSS1, KIAA0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase 1) (Chondroitin sulfate synthase 1) (N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS3_HUMAN
|
||||||
| NC score | 0.984780 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
CGAT1_MOUSE
|
||||||
| NC score | 0.807741 (rank : 4) | θ value | 1.15895e-47 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BJQ9, Q3UZX6, Q8BWV9, Q8BZU7, Q8C195, Q8R0M6 | Gene names | Galnact1, Chgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT1_HUMAN
|
||||||
| NC score | 0.805999 (rank : 5) | θ value | 2.3352e-48 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TDX6, Q6P9G6, Q8IUF9, Q9NSQ7, Q9NUM9 | Gene names | GALNACT1, CHGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT2_HUMAN
|
||||||
| NC score | 0.804713 (rank : 6) | θ value | 9.18288e-45 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N6G5, Q6MZJ5, Q6MZP6, Q8TCH4, Q9P1I6 | Gene names | GALNACT2, CHGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CGAT2_MOUSE
|
||||||
| NC score | 0.804179 (rank : 7) | θ value | 2.67181e-44 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8C1F4, Q6PAP6, Q8R5A2, Q9D2M1 | Gene names | Galnact2, Chgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CHSS2_MOUSE
|
||||||
| NC score | 0.716271 (rank : 8) | θ value | 3.05695e-40 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6IQX7 | Gene names | Css2, D1Bwg1363e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase). | |||||
|
CHSS2_HUMAN
|
||||||
| NC score | 0.711708 (rank : 9) | θ value | 3.4976e-36 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZ52, Q6UXD6, Q7L4G1, Q9H0F8, Q9H618 | Gene names | CSS2, CHPF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase) (Chondroitin-polymerizing factor). | |||||
|
CHGUT_HUMAN
|
||||||
| NC score | 0.710797 (rank : 10) | θ value | 1.98606e-31 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2E5, Q6P2I4, Q6UXD2 | Gene names | CSGLCAT, KIAA1402 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate glucuronyltransferase (EC 2.4.1.226) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (Chondroitin glucuronyltransferase II) (CSGlcA-T). | |||||
|
B4GN4_HUMAN
|
||||||
| NC score | 0.489091 (rank : 11) | θ value | 2.69671e-12 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GN4_MOUSE
|
||||||
| NC score | 0.488152 (rank : 12) | θ value | 6.00763e-12 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GN3_MOUSE
|
||||||
| NC score | 0.483064 (rank : 13) | θ value | 2.0648e-12 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6L8S8 | Gene names | B4galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
B4GN3_HUMAN
|
||||||
| NC score | 0.477405 (rank : 14) | θ value | 1.58096e-12 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6L9W6, Q6ZNC1, Q8N7T6 | Gene names | B4GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
B3GLT_MOUSE
|
||||||
| NC score | 0.401227 (rank : 15) | θ value | 5.43371e-13 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHT6, Q0PCR7, Q149S5 | Gene names | B3galtl, Gm1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
B3GLT_HUMAN
|
||||||
| NC score | 0.386581 (rank : 16) | θ value | 3.89403e-11 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6Y288, Q5W0H2, Q6NUI3 | Gene names | B3GALTL, B3GTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
RFNG_MOUSE
|
||||||
| NC score | 0.268525 (rank : 17) | θ value | 7.1131e-05 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09009 | Gene names | Rfng | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase radical fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
LFNG_MOUSE
|
||||||
| NC score | 0.261832 (rank : 18) | θ value | 2.21117e-06 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O09010, Q3U659, Q8K3F1, Q9DC10 | Gene names | Lfng | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase lunatic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
LFNG_HUMAN
|
||||||
| NC score | 0.248611 (rank : 19) | θ value | 1.09739e-05 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NES3, O00589, Q96C39, Q9UJW5 | Gene names | LFNG | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase lunatic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
RFNG_HUMAN
|
||||||
| NC score | 0.244470 (rank : 20) | θ value | 1.09739e-05 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y644, O00588 | Gene names | RFNG | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase radical fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
MFNG_MOUSE
|
||||||
| NC score | 0.198671 (rank : 21) | θ value | 0.0961366 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09008 | Gene names | Mfng | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase manic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
B4GT2_HUMAN
|
||||||
| NC score | 0.192731 (rank : 22) | θ value | 0.000121331 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60909, O60511, Q5T4X5, Q5T4Y5, Q9BUP6, Q9NSY7 | Gene names | B4GALT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT2_MOUSE
|
||||||
| NC score | 0.191344 (rank : 23) | θ value | 0.000461057 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Z2Y2, Q3TMP2 | Gene names | B4galt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
MFNG_HUMAN
|
||||||
| NC score | 0.178517 (rank : 24) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00587, O43730 | Gene names | MFNG | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase manic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
B4GT1_MOUSE
|
||||||
| NC score | 0.162421 (rank : 25) | θ value | 0.0193708 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P15535 | Gene names | B4galt1, Ggtb, Ggtb2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT1_HUMAN
|
||||||
| NC score | 0.151515 (rank : 26) | θ value | 0.0252991 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT4_MOUSE
|
||||||
| NC score | 0.138717 (rank : 27) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JJ04, Q8BR54, Q9QY12 | Gene names | B4galt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT4_HUMAN
|
||||||
| NC score | 0.134953 (rank : 28) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60513, Q68D68, Q9BSW3, Q9C078 | Gene names | B4GALT4 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT6_HUMAN
|
||||||
| NC score | 0.113500 (rank : 29) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBX8, O60514 | Gene names | B4GALT6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT6_MOUSE
|
||||||
| NC score | 0.113266 (rank : 30) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WVK5 | Gene names | B4galt6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT3_MOUSE
|
||||||
| NC score | 0.112538 (rank : 31) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91YY2, Q9QY13 | Gene names | B4galt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT3_HUMAN
|
||||||
| NC score | 0.111615 (rank : 32) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60512, O60910, Q9BPZ4, Q9H8T2 | Gene names | B4GALT3 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT7_MOUSE
|
||||||
| NC score | 0.111468 (rank : 33) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R087, Q7TNU3 | Gene names | B4galt7, Xgalt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
B4GT5_MOUSE
|
||||||
| NC score | 0.109864 (rank : 34) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JMK0 | Gene names | B4galt5, Bgt-5 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT5_HUMAN
|
||||||
| NC score | 0.109408 (rank : 35) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O43286, Q2M394, Q9UJQ8 | Gene names | B4GALT5 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT7_HUMAN
|
||||||
| NC score | 0.109019 (rank : 36) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UBV7, Q9UHN2 | Gene names | B4GALT7, XGALT1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
HRB2_HUMAN
|
||||||
| NC score | 0.049711 (rank : 37) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13601, Q8NEA8, Q8TC37, Q96AT5 | Gene names | HRB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV-1 Rev-binding protein 2 (Rev-interacting protein 1) (Rip-1). | |||||
|
HRB2_MOUSE
|
||||||
| NC score | 0.048351 (rank : 38) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGA5, Q8C029 | Gene names | Hrb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV-1 Rev-binding protein 2 homolog. | |||||
|
B3GL1_HUMAN
|
||||||
| NC score | 0.048218 (rank : 39) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75752, Q3Y531, Q6IAI5, Q8NFM8, Q9HA06 | Gene names | B3GALNT1, B3GALT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.79) (Beta-3-GalNAc-T1) (Beta-1,3-galactosyltransferase 3) (Beta-1,3- GalTase 3) (Beta3Gal-T3) (b3Gal-T3) (Galactosylgalactosylglucosylceramide beta-D-acetyl- galactosaminyltransferase) (UDP-N- acetylgalactosamine:globotriaosylceramide beta-1,3-N- acetylgalactosaminyltransferase) (Globoside synthase) (Beta-3-Gx-T3). | |||||
|
B3GL1_MOUSE
|
||||||
| NC score | 0.029297 (rank : 40) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q920V1, O54906, Q91V72, Q920V0, Q9CTE5 | Gene names | b3galnt1, B3galt3, B3gt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.79) (Beta-3-GalNAc-T1) (Beta-1,3-galactosyltransferase 3) (Beta-1,3- GalTase 3) (Beta3Gal-T3) (b3Gal-T3) (Galactosylgalactosylglucosylceramide beta-D-acetyl- galactosaminyltransferase) (UDP-N- acetylgalactosamine:globotriaosylceramide beta-1,3-N- acetylgalactosaminyltransferase) (Globoside synthase) (Beta-3-Gx-T3). | |||||
|
AHTF1_MOUSE
|
||||||
| NC score | 0.028214 (rank : 41) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CJF7, Q8BVJ5, Q8VD55 | Gene names | Ahctf1, Elys | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
NUDT6_HUMAN
|
||||||
| NC score | 0.024088 (rank : 42) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53370, O95097, Q9UQD9 | Gene names | NUDT6, FGF2AS | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 6 (Nudix motif 6) (Protein GFG) (GFG-1) (Antisense basic fibroblast growth factor). | |||||
|
FA26B_MOUSE
|
||||||
| NC score | 0.023522 (rank : 43) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VEC4, Q3TDQ7, Q8BT85, Q8C246 | Gene names | Fam26b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM26B. | |||||
|
CCD26_HUMAN
|
||||||
| NC score | 0.023314 (rank : 44) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TAB7 | Gene names | CCDC26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 26. | |||||
|
FA26B_HUMAN
|
||||||
| NC score | 0.020608 (rank : 45) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HA72, O95893, Q6ZUV9 | Gene names | FAM26B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM26B. | |||||
|
DDAH1_MOUSE
|
||||||
| NC score | 0.019975 (rank : 46) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CWS0, Q3UF65 | Gene names | Ddah1 | |||
|
Domain Architecture |

|
|||||
| Description | NG,NG-dimethylarginine dimethylaminohydrolase 1 (EC 3.5.3.18) (Dimethylargininase-1) (Dimethylarginine dimethylaminohydrolase 1) (DDAHI) (DDAH-1). | |||||
|
CCDC9_MOUSE
|
||||||
| NC score | 0.018564 (rank : 47) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
B3GT5_HUMAN
|
||||||
| NC score | 0.018491 (rank : 48) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2C3, Q9NY96, Q9P1X6, Q9P1X7 | Gene names | B3GALT5 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,3-GalTase 5) (Beta3Gal-T5) (b3Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,3-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,3- galactosyltransferase 5) (Beta-3-Gx-T5). | |||||
|
DDAH1_HUMAN
|
||||||
| NC score | 0.017626 (rank : 49) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94760, Q86XK5 | Gene names | DDAH1, DDAH | |||
|
Domain Architecture |

|
|||||
| Description | NG,NG-dimethylarginine dimethylaminohydrolase 1 (EC 3.5.3.18) (Dimethylargininase-1) (Dimethylarginine dimethylaminohydrolase 1) (DDAHI) (DDAH-1). | |||||
|
TRXR2_HUMAN
|
||||||
| NC score | 0.005918 (rank : 50) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NNW7, O95840, Q96IJ2, Q9H2Z5, Q9NZV3, Q9NZV4, Q9P2Y0, Q9P2Y1, Q9UQU8 | Gene names | TXNRD2, TRXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin reductase 2, mitochondrial precursor (EC 1.8.1.9) (TR3) (TR-beta) (Selenoprotein Z) (SelZ). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.004643 (rank : 51) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
LUZP1_HUMAN
|
||||||
| NC score | 0.003619 (rank : 52) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.003598 (rank : 53) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
TBX18_MOUSE
|
||||||
| NC score | 0.003047 (rank : 54) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPZ6 | Gene names | Tbx18 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
EMR1_MOUSE
|
||||||
| NC score | 0.001539 (rank : 55) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61549 | Gene names | Emr1, Gpf480 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 1 precursor (Cell surface glycoprotein EMR1) (EMR1 hormone receptor) (Cell surface glycoprotein F4/80). | |||||
|
RBCC1_MOUSE
|
||||||
| NC score | 0.000194 (rank : 56) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||