Please be patient as the page loads
|
B4GN3_MOUSE
|
||||||
| SwissProt Accessions | Q6L8S8 | Gene names | B4galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
B4GN3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.977199 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6L9W6, Q6ZNC1, Q8N7T6 | Gene names | B4GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
B4GN3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6L8S8 | Gene names | B4galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
B4GN4_HUMAN
|
||||||
| θ value | 6.86991e-125 (rank : 3) | NC score | 0.903593 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GN4_MOUSE
|
||||||
| θ value | 1.99884e-124 (rank : 4) | NC score | 0.925262 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
CGAT1_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 5) | NC score | 0.597116 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TDX6, Q6P9G6, Q8IUF9, Q9NSQ7, Q9NUM9 | Gene names | GALNACT1, CHGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT1_MOUSE
|
||||||
| θ value | 1.47631e-18 (rank : 6) | NC score | 0.594350 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BJQ9, Q3UZX6, Q8BWV9, Q8BZU7, Q8C195, Q8R0M6 | Gene names | Galnact1, Chgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT2_HUMAN
|
||||||
| θ value | 1.52774e-15 (rank : 7) | NC score | 0.577507 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N6G5, Q6MZJ5, Q6MZP6, Q8TCH4, Q9P1I6 | Gene names | GALNACT2, CHGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CGAT2_MOUSE
|
||||||
| θ value | 7.58209e-15 (rank : 8) | NC score | 0.571361 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C1F4, Q6PAP6, Q8R5A2, Q9D2M1 | Gene names | Galnact2, Chgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CHSS1_MOUSE
|
||||||
| θ value | 2.0648e-12 (rank : 9) | NC score | 0.483064 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS1_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 10) | NC score | 0.483834 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86X52, Q6UX38, Q7LFU5, Q9Y2J5 | Gene names | CHSY1, CHSY, CSS1, KIAA0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase 1) (Chondroitin sulfate synthase 1) (N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS3_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 11) | NC score | 0.466130 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
ROBO2_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 12) | NC score | 0.015716 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TPD3, Q8BJ59 | Gene names | Robo2, Kiaa1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
IF4G3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.047454 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
IF4G3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.047038 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.029598 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ST5_HUMAN
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.030353 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78524, P78523, Q16492, Q7KYY2, Q7KZ12, Q8NE12, Q9BQQ6 | Gene names | ST5, DENND2B, HTS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (HeLa tumor suppression 1) (DENN domain-containing protein 2B). | |||||
|
ST5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.027895 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q924W7, Q78H54, Q8K2P3, Q924W8 | Gene names | St5, Dennd2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (DENN domain-containing protein 2B). | |||||
|
RT10_MOUSE
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.037077 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80ZK0, Q4V9W5 | Gene names | Mrps10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial 28S ribosomal protein S10 (S10mt) (MRP-S10). | |||||
|
M4K4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.000423 (rank : 54) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
NIPA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.026971 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80YV2, Q3TJE6, Q80Z11, Q8BTW5, Q8CI56, Q8R3U7 | Gene names | Zc3hc1, Nipa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear-interacting partner of ALK (Nuclear-interacting partner of anaplastic lymphoma kinase) (mNIPA) (Zinc finger C3HC-type protein 1). | |||||
|
RIN3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.019914 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
GALT3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.029609 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70419, Q80V55 | Gene names | Galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 3) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 3) (Polypeptide GalNAc transferase 3) (GalNAc-T3) (pp-GaNTase 3). | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.020132 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.010101 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
B4GT2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.070556 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60909, O60511, Q5T4X5, Q5T4Y5, Q9BUP6, Q9NSY7 | Gene names | B4GALT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.017501 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
GALT3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.027420 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14435, Q7Z476 | Gene names | GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 3) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 3) (Polypeptide GalNAc transferase 3) (GalNAc-T3) (pp-GaNTase 3). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.004001 (rank : 50) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
B4GT2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.069321 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z2Y2, Q3TMP2 | Gene names | B4galt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.005870 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
EAA5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.009571 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00341 | Gene names | SLC1A7, EAAT5 | |||
|
Domain Architecture |
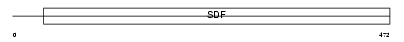
|
|||||
| Description | Excitatory amino acid transporter 5 (Retinal glutamate transporter). | |||||
|
GALT6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.026593 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NCL4, Q8IYH4, Q9H6G2, Q9UIV5 | Gene names | GALNT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
GP152_HUMAN
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.008211 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
GPR64_MOUSE
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.007622 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CJ12, Q8BL10, Q8CJ08, Q8CJ09, Q8CJ10 | Gene names | Gpr64, Me6 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (Me6 receptor). | |||||
|
LAP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.005069 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TH2, Q8BQ14, Q8CE41, Q8K171, Q99JU3, Q9JI47 | Gene names | Erbb2ip, Erbin, Kiaa1225, Lap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.021658 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
S4A11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.012368 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NBS3, Q9BXF4, Q9NTW9 | Gene names | SLC4A11, BTR1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium bicarbonate transporter-like protein 11 (Bicarbonate transporter-related protein 1) (Solute carrier family 4 member 11). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.040754 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.039145 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
VHL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.024056 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P40337, Q13599 | Gene names | VHL | |||
|
Domain Architecture |
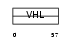
|
|||||
| Description | Von Hippel-Lindau disease tumor suppressor (pVHL) (G7 protein). | |||||
|
AP3B1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.008681 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
RASF4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.012472 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CB96, Q3TBK1, Q3TPH9, Q3TUT0, Q6P9L7, Q8CIM6 | Gene names | Rassf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras association domain-containing protein 4. | |||||
|
CGRE1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.015949 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
ZCHC9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.014661 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N567, Q9H027 | Gene names | ZCCHC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 9. | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.003306 (rank : 52) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.000890 (rank : 53) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
GALT6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.020598 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C7U7, Q3TWF0, Q8CED2, Q9QZ16 | Gene names | Galnt6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
LRCH4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.003980 (rank : 51) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921G6 | Gene names | Lrch4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4. | |||||
|
SYN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.007767 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
B3GLT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.052415 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6Y288, Q5W0H2, Q6NUI3 | Gene names | B3GALTL, B3GTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
B3GLT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.056640 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHT6, Q0PCR7, Q149S5 | Gene names | B3galtl, Gm1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
CHGUT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.152577 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P2E5, Q6P2I4, Q6UXD2 | Gene names | CSGLCAT, KIAA1402 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate glucuronyltransferase (EC 2.4.1.226) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (Chondroitin glucuronyltransferase II) (CSGlcA-T). | |||||
|
CHSS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.177322 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IZ52, Q6UXD6, Q7L4G1, Q9H0F8, Q9H618 | Gene names | CSS2, CHPF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase) (Chondroitin-polymerizing factor). | |||||
|
CHSS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.176934 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6IQX7 | Gene names | Css2, D1Bwg1363e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase). | |||||
|
B4GN3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6L8S8 | Gene names | B4galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
B4GN3_HUMAN
|
||||||
| NC score | 0.977199 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6L9W6, Q6ZNC1, Q8N7T6 | Gene names | B4GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
B4GN4_MOUSE
|
||||||
| NC score | 0.925262 (rank : 3) | θ value | 1.99884e-124 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GN4_HUMAN
|
||||||
| NC score | 0.903593 (rank : 4) | θ value | 6.86991e-125 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
CGAT1_HUMAN
|
||||||
| NC score | 0.597116 (rank : 5) | θ value | 5.07402e-19 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TDX6, Q6P9G6, Q8IUF9, Q9NSQ7, Q9NUM9 | Gene names | GALNACT1, CHGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT1_MOUSE
|
||||||
| NC score | 0.594350 (rank : 6) | θ value | 1.47631e-18 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BJQ9, Q3UZX6, Q8BWV9, Q8BZU7, Q8C195, Q8R0M6 | Gene names | Galnact1, Chgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT2_HUMAN
|
||||||
| NC score | 0.577507 (rank : 7) | θ value | 1.52774e-15 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N6G5, Q6MZJ5, Q6MZP6, Q8TCH4, Q9P1I6 | Gene names | GALNACT2, CHGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CGAT2_MOUSE
|
||||||
| NC score | 0.571361 (rank : 8) | θ value | 7.58209e-15 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C1F4, Q6PAP6, Q8R5A2, Q9D2M1 | Gene names | Galnact2, Chgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CHSS1_HUMAN
|
||||||
| NC score | 0.483834 (rank : 9) | θ value | 7.84624e-12 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86X52, Q6UX38, Q7LFU5, Q9Y2J5 | Gene names | CHSY1, CHSY, CSS1, KIAA0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase 1) (Chondroitin sulfate synthase 1) (N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS1_MOUSE
|
||||||
| NC score | 0.483064 (rank : 10) | θ value | 2.0648e-12 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS3_HUMAN
|
||||||
| NC score | 0.466130 (rank : 11) | θ value | 1.25267e-09 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
CHSS2_HUMAN
|
||||||
| NC score | 0.177322 (rank : 12) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IZ52, Q6UXD6, Q7L4G1, Q9H0F8, Q9H618 | Gene names | CSS2, CHPF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase) (Chondroitin-polymerizing factor). | |||||
|
CHSS2_MOUSE
|
||||||
| NC score | 0.176934 (rank : 13) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6IQX7 | Gene names | Css2, D1Bwg1363e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase). | |||||
|
CHGUT_HUMAN
|
||||||
| NC score | 0.152577 (rank : 14) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P2E5, Q6P2I4, Q6UXD2 | Gene names | CSGLCAT, KIAA1402 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate glucuronyltransferase (EC 2.4.1.226) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (Chondroitin glucuronyltransferase II) (CSGlcA-T). | |||||
|
B4GT2_HUMAN
|
||||||
| NC score | 0.070556 (rank : 15) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60909, O60511, Q5T4X5, Q5T4Y5, Q9BUP6, Q9NSY7 | Gene names | B4GALT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT2_MOUSE
|
||||||
| NC score | 0.069321 (rank : 16) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z2Y2, Q3TMP2 | Gene names | B4galt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B3GLT_MOUSE
|
||||||
| NC score | 0.056640 (rank : 17) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHT6, Q0PCR7, Q149S5 | Gene names | B3galtl, Gm1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
B3GLT_HUMAN
|
||||||
| NC score | 0.052415 (rank : 18) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6Y288, Q5W0H2, Q6NUI3 | Gene names | B3GALTL, B3GTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
IF4G3_HUMAN
|
||||||
| NC score | 0.047454 (rank : 19) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
IF4G3_MOUSE
|
||||||
| NC score | 0.047038 (rank : 20) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.040754 (rank : 21) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.039145 (rank : 22) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
RT10_MOUSE
|
||||||
| NC score | 0.037077 (rank : 23) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80ZK0, Q4V9W5 | Gene names | Mrps10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial 28S ribosomal protein S10 (S10mt) (MRP-S10). | |||||
|
ST5_HUMAN
|
||||||
| NC score | 0.030353 (rank : 24) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78524, P78523, Q16492, Q7KYY2, Q7KZ12, Q8NE12, Q9BQQ6 | Gene names | ST5, DENND2B, HTS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (HeLa tumor suppression 1) (DENN domain-containing protein 2B). | |||||
|
GALT3_MOUSE
|
||||||
| NC score | 0.029609 (rank : 25) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70419, Q80V55 | Gene names | Galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 3) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 3) (Polypeptide GalNAc transferase 3) (GalNAc-T3) (pp-GaNTase 3). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.029598 (rank : 26) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ST5_MOUSE
|
||||||
| NC score | 0.027895 (rank : 27) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q924W7, Q78H54, Q8K2P3, Q924W8 | Gene names | St5, Dennd2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity 5 (DENN domain-containing protein 2B). | |||||
|
GALT3_HUMAN
|
||||||
| NC score | 0.027420 (rank : 28) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14435, Q7Z476 | Gene names | GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 3) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 3) (Polypeptide GalNAc transferase 3) (GalNAc-T3) (pp-GaNTase 3). | |||||
|
NIPA_MOUSE
|
||||||
| NC score | 0.026971 (rank : 29) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80YV2, Q3TJE6, Q80Z11, Q8BTW5, Q8CI56, Q8R3U7 | Gene names | Zc3hc1, Nipa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear-interacting partner of ALK (Nuclear-interacting partner of anaplastic lymphoma kinase) (mNIPA) (Zinc finger C3HC-type protein 1). | |||||
|
GALT6_HUMAN
|
||||||
| NC score | 0.026593 (rank : 30) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NCL4, Q8IYH4, Q9H6G2, Q9UIV5 | Gene names | GALNT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
VHL_HUMAN
|
||||||
| NC score | 0.024056 (rank : 31) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P40337, Q13599 | Gene names | VHL | |||
|
Domain Architecture |

|
|||||
| Description | Von Hippel-Lindau disease tumor suppressor (pVHL) (G7 protein). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.021658 (rank : 32) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
GALT6_MOUSE
|
||||||
| NC score | 0.020598 (rank : 33) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C7U7, Q3TWF0, Q8CED2, Q9QZ16 | Gene names | Galnt6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.020132 (rank : 34) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
RIN3_MOUSE
|
||||||
| NC score | 0.019914 (rank : 35) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.017501 (rank : 36) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CGRE1_MOUSE
|
||||||
| NC score | 0.015949 (rank : 37) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
ROBO2_MOUSE
|
||||||
| NC score | 0.015716 (rank : 38) | θ value | 0.0148317 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TPD3, Q8BJ59 | Gene names | Robo2, Kiaa1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
ZCHC9_HUMAN
|
||||||
| NC score | 0.014661 (rank : 39) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N567, Q9H027 | Gene names | ZCCHC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 9. | |||||
|
RASF4_MOUSE
|
||||||
| NC score | 0.012472 (rank : 40) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CB96, Q3TBK1, Q3TPH9, Q3TUT0, Q6P9L7, Q8CIM6 | Gene names | Rassf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras association domain-containing protein 4. | |||||
|
S4A11_HUMAN
|
||||||
| NC score | 0.012368 (rank : 41) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NBS3, Q9BXF4, Q9NTW9 | Gene names | SLC4A11, BTR1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium bicarbonate transporter-like protein 11 (Bicarbonate transporter-related protein 1) (Solute carrier family 4 member 11). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.010101 (rank : 42) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
EAA5_HUMAN
|
||||||
| NC score | 0.009571 (rank : 43) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00341 | Gene names | SLC1A7, EAAT5 | |||
|
Domain Architecture |

|
|||||
| Description | Excitatory amino acid transporter 5 (Retinal glutamate transporter). | |||||
|
AP3B1_HUMAN
|
||||||
| NC score | 0.008681 (rank : 44) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
GP152_HUMAN
|
||||||
| NC score | 0.008211 (rank : 45) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.007767 (rank : 46) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
GPR64_MOUSE
|
||||||
| NC score | 0.007622 (rank : 47) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CJ12, Q8BL10, Q8CJ08, Q8CJ09, Q8CJ10 | Gene names | Gpr64, Me6 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (Me6 receptor). | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.005870 (rank : 48) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
LAP2_MOUSE
|
||||||
| NC score | 0.005069 (rank : 49) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TH2, Q8BQ14, Q8CE41, Q8K171, Q99JU3, Q9JI47 | Gene names | Erbb2ip, Erbin, Kiaa1225, Lap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.004001 (rank : 50) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
LRCH4_MOUSE
|
||||||
| NC score | 0.003980 (rank : 51) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921G6 | Gene names | Lrch4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4. | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.003306 (rank : 52) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.000890 (rank : 53) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
M4K4_HUMAN
|
||||||
| NC score | 0.000423 (rank : 54) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||