Please be patient as the page loads
|
CHGUT_HUMAN
|
||||||
| SwissProt Accessions | Q9P2E5, Q6P2I4, Q6UXD2 | Gene names | CSGLCAT, KIAA1402 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate glucuronyltransferase (EC 2.4.1.226) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (Chondroitin glucuronyltransferase II) (CSGlcA-T). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CHGUT_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9P2E5, Q6P2I4, Q6UXD2 | Gene names | CSGLCAT, KIAA1402 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate glucuronyltransferase (EC 2.4.1.226) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (Chondroitin glucuronyltransferase II) (CSGlcA-T). | |||||
|
CHSS2_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.978275 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IZ52, Q6UXD6, Q7L4G1, Q9H0F8, Q9H618 | Gene names | CSS2, CHPF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase) (Chondroitin-polymerizing factor). | |||||
|
CHSS2_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.979104 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6IQX7 | Gene names | Css2, D1Bwg1363e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase). | |||||
|
CHSS3_HUMAN
|
||||||
| θ value | 1.28434e-38 (rank : 4) | NC score | 0.718281 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
CHSS1_HUMAN
|
||||||
| θ value | 1.98606e-31 (rank : 5) | NC score | 0.710660 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86X52, Q6UX38, Q7LFU5, Q9Y2J5 | Gene names | CHSY1, CHSY, CSS1, KIAA0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase 1) (Chondroitin sulfate synthase 1) (N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS1_MOUSE
|
||||||
| θ value | 1.98606e-31 (rank : 6) | NC score | 0.710797 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CGAT2_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 7) | NC score | 0.476460 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N6G5, Q6MZJ5, Q6MZP6, Q8TCH4, Q9P1I6 | Gene names | GALNACT2, CHGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CGAT2_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 8) | NC score | 0.475924 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C1F4, Q6PAP6, Q8R5A2, Q9D2M1 | Gene names | Galnact2, Chgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CGAT1_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 9) | NC score | 0.460663 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TDX6, Q6P9G6, Q8IUF9, Q9NSQ7, Q9NUM9 | Gene names | GALNACT1, CHGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
RFNG_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 10) | NC score | 0.213235 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09009 | Gene names | Rfng | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase radical fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
CGAT1_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 11) | NC score | 0.462973 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BJQ9, Q3UZX6, Q8BWV9, Q8BZU7, Q8C195, Q8R0M6 | Gene names | Galnact1, Chgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
RFNG_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 12) | NC score | 0.183669 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y644, O00588 | Gene names | RFNG | |||
|
Domain Architecture |
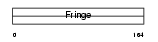
|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase radical fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
MFNG_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 13) | NC score | 0.144751 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00587, O43730 | Gene names | MFNG | |||
|
Domain Architecture |
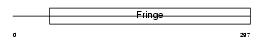
|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase manic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 0.125558 (rank : 14) | NC score | 0.019529 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
LFNG_MOUSE
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.179639 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09010, Q3U659, Q8K3F1, Q9DC10 | Gene names | Lfng | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase lunatic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
B4GN4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.196042 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
LFNG_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.169449 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NES3, O00589, Q96C39, Q9UJW5 | Gene names | LFNG | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase lunatic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
B3GLT_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.192484 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6Y288, Q5W0H2, Q6NUI3 | Gene names | B3GALTL, B3GTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
B3GLT_MOUSE
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.200168 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHT6, Q0PCR7, Q149S5 | Gene names | B3galtl, Gm1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
PON2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.031747 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15165, O15114, O15115, O75856, Q86YL0 | Gene names | PON2 | |||
|
Domain Architecture |

|
|||||
| Description | Serum paraoxonase/arylesterase 2 (EC 3.1.1.2) (EC 3.1.8.1) (PON 2) (Serum aryldialkylphosphatase 2) (A-esterase 2) (Aromatic esterase 2). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.018570 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
PON2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.028904 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62086, Q8CFK3 | Gene names | Pon2 | |||
|
Domain Architecture |

|
|||||
| Description | Serum paraoxonase/arylesterase 2 (EC 3.1.1.2) (EC 3.1.8.1) (PON 2) (Serum aryldialkylphosphatase 2) (A-esterase 2) (Aromatic esterase 2). | |||||
|
MFNG_MOUSE
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.143954 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09008 | Gene names | Mfng | |||
|
Domain Architecture |
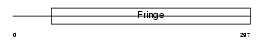
|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase manic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
B4GN3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.157892 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6L9W6, Q6ZNC1, Q8N7T6 | Gene names | B4GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
BT1A1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.006359 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13410, Q9H458 | Gene names | BTN1A1, BTN | |||
|
Domain Architecture |

|
|||||
| Description | Butyrophilin subfamily 1 member A1 precursor (BT). | |||||
|
KIF11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.003591 (rank : 35) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52732, Q15716, Q5VWX0 | Gene names | KIF11, EG5, KNSL1 | |||
|
Domain Architecture |
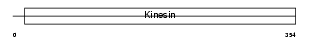
|
|||||
| Description | Kinesin-like protein KIF11 (Kinesin-related motor protein Eg5) (Kinesin-like spindle protein HKSP) (Thyroid receptor-interacting protein 5) (TRIP5) (Kinesin-like protein 1). | |||||
|
LYAG_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.010345 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10253, Q14351, Q16302 | Gene names | GAA | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-glucosidase precursor (EC 3.2.1.20) (Acid maltase) (Aglucosidase alfa) [Contains: 76 kDa lysosomal alpha-glucosidase; 70 kDa lysosomal alpha-glucosidase]. | |||||
|
BT1A1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.006082 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62556, P97392, Q8K2H7, Q921K7 | Gene names | Btn1a1, Btn | |||
|
Domain Architecture |

|
|||||
| Description | Butyrophilin subfamily 1 member A1 precursor (BT). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.003533 (rank : 36) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PSMF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.015443 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BHL8, Q8C0G9 | Gene names | Psmf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome inhibitor PI31 subunit. | |||||
|
SMBT2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.008691 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5DTW2, Q80VG7 | Gene names | Sfmbt2, Kiaa1617 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 2. | |||||
|
ANGL7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.003863 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R1Q3 | Gene names | Angptl7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 7 precursor (Angiopoietin-like 7). | |||||
|
SAPS2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.007231 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75170, Q9UGB9 | Gene names | SAPS2, KIAA0685 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 2. | |||||
|
TNAP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.009125 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P21580, Q5VXQ7, Q9NSR6 | Gene names | TNFAIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 3 (EC 3.-.-.-) (Putative DNA-binding protein A20) (Zinc finger protein A20). | |||||
|
B4GN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.152577 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6L8S8 | Gene names | B4galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
B4GN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.190445 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
CHGUT_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9P2E5, Q6P2I4, Q6UXD2 | Gene names | CSGLCAT, KIAA1402 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate glucuronyltransferase (EC 2.4.1.226) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (Chondroitin glucuronyltransferase II) (CSGlcA-T). | |||||
|
CHSS2_MOUSE
|
||||||
| NC score | 0.979104 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6IQX7 | Gene names | Css2, D1Bwg1363e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase). | |||||
|
CHSS2_HUMAN
|
||||||
| NC score | 0.978275 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IZ52, Q6UXD6, Q7L4G1, Q9H0F8, Q9H618 | Gene names | CSS2, CHPF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase) (Chondroitin-polymerizing factor). | |||||
|
CHSS3_HUMAN
|
||||||
| NC score | 0.718281 (rank : 4) | θ value | 1.28434e-38 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
CHSS1_MOUSE
|
||||||
| NC score | 0.710797 (rank : 5) | θ value | 1.98606e-31 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS1_HUMAN
|
||||||
| NC score | 0.710660 (rank : 6) | θ value | 1.98606e-31 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86X52, Q6UX38, Q7LFU5, Q9Y2J5 | Gene names | CHSY1, CHSY, CSS1, KIAA0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase 1) (Chondroitin sulfate synthase 1) (N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CGAT2_HUMAN
|
||||||
| NC score | 0.476460 (rank : 7) | θ value | 7.59969e-07 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8N6G5, Q6MZJ5, Q6MZP6, Q8TCH4, Q9P1I6 | Gene names | GALNACT2, CHGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CGAT2_MOUSE
|
||||||
| NC score | 0.475924 (rank : 8) | θ value | 7.59969e-07 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C1F4, Q6PAP6, Q8R5A2, Q9D2M1 | Gene names | Galnact2, Chgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CGAT1_MOUSE
|
||||||
| NC score | 0.462973 (rank : 9) | θ value | 1.87187e-05 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BJQ9, Q3UZX6, Q8BWV9, Q8BZU7, Q8C195, Q8R0M6 | Gene names | Galnact1, Chgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT1_HUMAN
|
||||||
| NC score | 0.460663 (rank : 10) | θ value | 3.77169e-06 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TDX6, Q6P9G6, Q8IUF9, Q9NSQ7, Q9NUM9 | Gene names | GALNACT1, CHGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
RFNG_MOUSE
|
||||||
| NC score | 0.213235 (rank : 11) | θ value | 3.77169e-06 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09009 | Gene names | Rfng | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase radical fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
B3GLT_MOUSE
|
||||||
| NC score | 0.200168 (rank : 12) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHT6, Q0PCR7, Q149S5 | Gene names | B3galtl, Gm1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
B4GN4_MOUSE
|
||||||
| NC score | 0.196042 (rank : 13) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B3GLT_HUMAN
|
||||||
| NC score | 0.192484 (rank : 14) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6Y288, Q5W0H2, Q6NUI3 | Gene names | B3GALTL, B3GTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
B4GN4_HUMAN
|
||||||
| NC score | 0.190445 (rank : 15) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
RFNG_HUMAN
|
||||||
| NC score | 0.183669 (rank : 16) | θ value | 0.00869519 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y644, O00588 | Gene names | RFNG | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase radical fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
LFNG_MOUSE
|
||||||
| NC score | 0.179639 (rank : 17) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09010, Q3U659, Q8K3F1, Q9DC10 | Gene names | Lfng | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase lunatic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
LFNG_HUMAN
|
||||||
| NC score | 0.169449 (rank : 18) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NES3, O00589, Q96C39, Q9UJW5 | Gene names | LFNG | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase lunatic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
B4GN3_HUMAN
|
||||||
| NC score | 0.157892 (rank : 19) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6L9W6, Q6ZNC1, Q8N7T6 | Gene names | B4GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
B4GN3_MOUSE
|
||||||
| NC score | 0.152577 (rank : 20) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6L8S8 | Gene names | B4galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
MFNG_HUMAN
|
||||||
| NC score | 0.144751 (rank : 21) | θ value | 0.0736092 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00587, O43730 | Gene names | MFNG | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase manic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
MFNG_MOUSE
|
||||||
| NC score | 0.143954 (rank : 22) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09008 | Gene names | Mfng | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase manic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
PON2_HUMAN
|
||||||
| NC score | 0.031747 (rank : 23) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15165, O15114, O15115, O75856, Q86YL0 | Gene names | PON2 | |||
|
Domain Architecture |

|
|||||
| Description | Serum paraoxonase/arylesterase 2 (EC 3.1.1.2) (EC 3.1.8.1) (PON 2) (Serum aryldialkylphosphatase 2) (A-esterase 2) (Aromatic esterase 2). | |||||
|
PON2_MOUSE
|
||||||
| NC score | 0.028904 (rank : 24) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62086, Q8CFK3 | Gene names | Pon2 | |||
|
Domain Architecture |

|
|||||
| Description | Serum paraoxonase/arylesterase 2 (EC 3.1.1.2) (EC 3.1.8.1) (PON 2) (Serum aryldialkylphosphatase 2) (A-esterase 2) (Aromatic esterase 2). | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.019529 (rank : 25) | θ value | 0.125558 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.018570 (rank : 26) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
PSMF1_MOUSE
|
||||||
| NC score | 0.015443 (rank : 27) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BHL8, Q8C0G9 | Gene names | Psmf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome inhibitor PI31 subunit. | |||||
|
LYAG_HUMAN
|
||||||
| NC score | 0.010345 (rank : 28) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10253, Q14351, Q16302 | Gene names | GAA | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-glucosidase precursor (EC 3.2.1.20) (Acid maltase) (Aglucosidase alfa) [Contains: 76 kDa lysosomal alpha-glucosidase; 70 kDa lysosomal alpha-glucosidase]. | |||||
|
TNAP3_HUMAN
|
||||||
| NC score | 0.009125 (rank : 29) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P21580, Q5VXQ7, Q9NSR6 | Gene names | TNFAIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 3 (EC 3.-.-.-) (Putative DNA-binding protein A20) (Zinc finger protein A20). | |||||
|
SMBT2_MOUSE
|
||||||
| NC score | 0.008691 (rank : 30) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5DTW2, Q80VG7 | Gene names | Sfmbt2, Kiaa1617 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 2. | |||||
|
SAPS2_HUMAN
|
||||||
| NC score | 0.007231 (rank : 31) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75170, Q9UGB9 | Gene names | SAPS2, KIAA0685 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 2. | |||||
|
BT1A1_HUMAN
|
||||||
| NC score | 0.006359 (rank : 32) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13410, Q9H458 | Gene names | BTN1A1, BTN | |||
|
Domain Architecture |

|
|||||
| Description | Butyrophilin subfamily 1 member A1 precursor (BT). | |||||
|
BT1A1_MOUSE
|
||||||
| NC score | 0.006082 (rank : 33) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62556, P97392, Q8K2H7, Q921K7 | Gene names | Btn1a1, Btn | |||
|
Domain Architecture |

|
|||||
| Description | Butyrophilin subfamily 1 member A1 precursor (BT). | |||||
|
ANGL7_MOUSE
|
||||||
| NC score | 0.003863 (rank : 34) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R1Q3 | Gene names | Angptl7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 7 precursor (Angiopoietin-like 7). | |||||
|
KIF11_HUMAN
|
||||||
| NC score | 0.003591 (rank : 35) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52732, Q15716, Q5VWX0 | Gene names | KIF11, EG5, KNSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF11 (Kinesin-related motor protein Eg5) (Kinesin-like spindle protein HKSP) (Thyroid receptor-interacting protein 5) (TRIP5) (Kinesin-like protein 1). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.003533 (rank : 36) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||