Please be patient as the page loads
|
PSMF1_MOUSE
|
||||||
| SwissProt Accessions | Q8BHL8, Q8C0G9 | Gene names | Psmf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome inhibitor PI31 subunit. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PSMF1_MOUSE
|
||||||
| θ value | 3.4876e-161 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8BHL8, Q8C0G9 | Gene names | Psmf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome inhibitor PI31 subunit. | |||||
|
PSMF1_HUMAN
|
||||||
| θ value | 2.97304e-136 (rank : 2) | NC score | 0.958415 (rank : 2) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92530, Q9H4I1 | Gene names | PSMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome inhibitor PI31 subunit (hPI31). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 3) | NC score | 0.114719 (rank : 7) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 4) | NC score | 0.122880 (rank : 3) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 5) | NC score | 0.120600 (rank : 4) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
CPSF6_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 6) | NC score | 0.116210 (rank : 5) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CPSF6_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 7) | NC score | 0.115578 (rank : 6) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 8) | NC score | 0.019780 (rank : 76) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 9) | NC score | 0.052354 (rank : 30) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
PB1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 10) | NC score | 0.047178 (rank : 38) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.041923 (rank : 39) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SF3B4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.037271 (rank : 47) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.060257 (rank : 20) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
CN032_MOUSE
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.058851 (rank : 23) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
RXRB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.022429 (rank : 67) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P28702, P28703 | Gene names | RXRB, NR2B2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-beta (Retinoid X receptor beta). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.051227 (rank : 34) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
DAZP1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.037306 (rank : 46) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96EP5, Q96MJ3, Q9NRR9 | Gene names | DAZAP1 | |||
|
Domain Architecture |
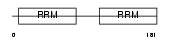
|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
DAZP1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.037222 (rank : 48) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JII5 | Gene names | Dazap1 | |||
|
Domain Architecture |
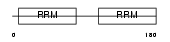
|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.058349 (rank : 24) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
ESX1L_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.013168 (rank : 93) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N693, Q7Z6K7 | Gene names | ESX1L, ESX1R | |||
|
Domain Architecture |

|
|||||
| Description | Extraembryonic, spermatogenesis, homeobox 1-like protein. | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.051550 (rank : 32) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
DYN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.036494 (rank : 52) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q05193 | Gene names | DNM1, DNM | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
DYN1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.036521 (rank : 51) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P39053, Q61358, Q61359, Q61360, Q8JZZ4, Q9CSY7, Q9QXX1 | Gene names | Dnm1, Dnm | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
FBX7_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.065763 (rank : 16) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y3I1, Q5TGC4, Q96HM6, Q9UF21, Q9UKT2 | Gene names | FBXO7, FBX7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 7. | |||||
|
NFIC_HUMAN
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.026505 (rank : 60) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08651, P08652, Q14932, Q9UPJ3, Q9UPJ9, Q9UPK0, Q9UPK1 | Gene names | NFIC, NFI | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 C-type (Nuclear factor 1/C) (NF1-C) (NFI-C) (NF-I/C) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
CS007_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.032129 (rank : 58) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
PP1RA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.048453 (rank : 37) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96QC0, O00405 | Gene names | PPP1R10, CAT53, FB19, PNUTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (Phosphatase 1 nuclear targeting subunit) (MHC class I region proline- rich protein CAT53) (FB19 protein) (PP1-binding protein of 114 kDa) (p99). | |||||
|
RSMN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.060044 (rank : 21) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P63162, P14648, P17135 | Gene names | SNRPN, HCERN3, SMN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
RSMN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.060044 (rank : 22) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P63163, P14648, P17135 | Gene names | Snrpn, Smn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
WASF3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.041243 (rank : 41) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VHI6 | Gene names | Wasf3, Wave3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3). | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.039278 (rank : 43) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
CO2A1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.020560 (rank : 75) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P28481 | Gene names | Col2a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
CPSF7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.092563 (rank : 8) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N684, Q7Z3H9, Q9H025, Q9H9V1 | Gene names | CPSF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7 (Cleavage and polyadenylation specificity factor 59 kDa subunit) (CPSF 59 kDa subunit) (Pre-mRNA cleavage factor Im 59 kDa subunit). | |||||
|
CPSF7_MOUSE
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.092240 (rank : 10) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BTV2, Q3TNF1, Q8BKE7, Q8CFS8 | Gene names | Cpsf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.020862 (rank : 74) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.023993 (rank : 64) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
WASF4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.040892 (rank : 42) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
WASL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.034650 (rank : 55) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
BAI1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.015621 (rank : 85) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
BT2A3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.010805 (rank : 96) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96KV6 | Gene names | BTN2A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Butyrophilin subfamily 2 member A3 precursor. | |||||
|
CN032_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.041770 (rank : 40) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.017239 (rank : 80) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
PRR13_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.080069 (rank : 11) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.030568 (rank : 59) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
TENS1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.025566 (rank : 62) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
WBP11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.038424 (rank : 44) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
3BP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.015741 (rank : 84) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
BAI1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.015104 (rank : 87) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3UHD1, Q3UH36, Q8CGM0 | Gene names | Bai1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
DAAM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.021145 (rank : 72) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BPM0, Q80Y68, Q9CQQ2 | Gene names | Daam1 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
EPN1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.033754 (rank : 57) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
SC24D_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.023102 (rank : 66) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94855 | Gene names | SEC24D, KIAA0755 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24D (SEC24-related protein D). | |||||
|
WASF2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.037803 (rank : 45) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
AATK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.010959 (rank : 95) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
G3BP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.024173 (rank : 63) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97855 | Gene names | G3bp, G3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 1 (EC 3.6.1.-) (ATP- dependent DNA helicase VIII) (GAP SH3-domain-binding protein 1) (G3BP- 1) (HDH-VIII). | |||||
|
IKBE_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.013388 (rank : 92) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54910, Q3U686, Q9CZZ9, Q9D7U3 | Gene names | Nfkbie, Ikbe | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKB-E). | |||||
|
MCM3A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.022216 (rank : 69) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WUU9 | Gene names | Mcm3ap, Ganp, Map80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
RXRB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.013983 (rank : 89) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28704, P33243 | Gene names | Rxrb, Nr2b2 | |||
|
Domain Architecture |
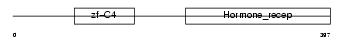
|
|||||
| Description | Retinoic acid receptor RXR-beta (Retinoid X receptor beta) (MHC class I regulatory element-binding protein H-2RIIBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.036296 (rank : 54) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.022395 (rank : 68) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.023432 (rank : 65) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
C1QRF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.013621 (rank : 91) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88992 | Gene names | C1ql1, C1qrf, Crf | |||
|
Domain Architecture |
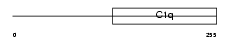
|
|||||
| Description | C1q-related factor precursor (Complement component 1 Q subcomponent- like 1). | |||||
|
CHGUT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.015443 (rank : 86) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P2E5, Q6P2I4, Q6UXD2 | Gene names | CSGLCAT, KIAA1402 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate glucuronyltransferase (EC 2.4.1.226) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (Chondroitin glucuronyltransferase II) (CSGlcA-T). | |||||
|
CO3A1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.013978 (rank : 90) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
FUBP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.021509 (rank : 70) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96AE4, Q12828 | Gene names | FUBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP) (DNA helicase V) (HDH V). | |||||
|
FUBP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.021421 (rank : 71) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91WJ8, Q8C0Y8 | Gene names | Fubp1, D3Ertd330e | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.016074 (rank : 83) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
SF01_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.017130 (rank : 81) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
TNFL6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.017712 (rank : 79) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41047, Q61217, Q9R1F2 | Gene names | Faslg, Apt1lg1, Fasl, gld, Tnfsf6 | |||
|
Domain Architecture |
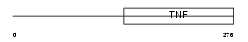
|
|||||
| Description | Tumor necrosis factor ligand superfamily member 6 (Fas antigen ligand) (Fas ligand) (CD178 antigen) (CD95L protein) [Contains: Tumor necrosis factor ligand superfamily member 6, membrane form; Tumor necrosis factor ligand superfamily member 6, soluble form]. | |||||
|
WASL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.034520 (rank : 56) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.037055 (rank : 49) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.036302 (rank : 53) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.058094 (rank : 25) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
COIA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.012347 (rank : 94) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
DOCK8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.009766 (rank : 97) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NF50, Q3MV16, Q5JPJ1, Q8TEP1, Q8WUY2, Q9BYJ5, Q9H1Q2, Q9H1Q3, Q9H308, Q9H7P2 | Gene names | DOCK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 8. | |||||
|
FMN1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.018800 (rank : 78) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q05860 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoforms I/II/III (Limb deformity protein). | |||||
|
FMN1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.019320 (rank : 77) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05859 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoform IV (Limb deformity protein). | |||||
|
MK07_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | -0.000322 (rank : 98) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.016193 (rank : 82) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PP1RA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.036635 (rank : 50) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80W00, Q811B6, Q8C6T7, Q8K2U8 | Gene names | Ppp1r10, Cat53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (MHC class I region proline-rich protein CAT53). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.013988 (rank : 88) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
RBM16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.025843 (rank : 61) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
RSMB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.050677 (rank : 35) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P14678, Q15490, Q9UIS5 | Gene names | SNRPB, COD, SNRPB1 | |||
|
Domain Architecture |
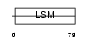
|
|||||
| Description | Small nuclear ribonucleoprotein-associated proteins B and B' (snRNP-B) (Sm protein B/B') (Sm-B/Sm-B') (SmB/SmB'). | |||||
|
RSMB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.050480 (rank : 36) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P27048, Q3UJT1 | Gene names | Snrpb | |||
|
Domain Architecture |
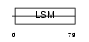
|
|||||
| Description | Small nuclear ribonucleoprotein-associated protein B (snRNP-B) (Sm protein B) (Sm-B) (SmB). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.054222 (rank : 26) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
ZN207_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.021028 (rank : 73) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
BCL9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.070084 (rank : 13) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CECR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.051604 (rank : 31) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
CEL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.066053 (rank : 15) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
MISS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.092258 (rank : 9) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.062796 (rank : 18) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.060331 (rank : 19) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.067585 (rank : 14) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SC24C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.063533 (rank : 17) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
SON_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.052402 (rank : 29) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.076347 (rank : 12) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
TPO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.052565 (rank : 28) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.052887 (rank : 27) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.051547 (rank : 33) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
PSMF1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 3.4876e-161 (rank : 1) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8BHL8, Q8C0G9 | Gene names | Psmf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome inhibitor PI31 subunit. | |||||
|
PSMF1_HUMAN
|
||||||
| NC score | 0.958415 (rank : 2) | θ value | 2.97304e-136 (rank : 2) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92530, Q9H4I1 | Gene names | PSMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome inhibitor PI31 subunit (hPI31). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.122880 (rank : 3) | θ value | 0.00869519 (rank : 4) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.120600 (rank : 4) | θ value | 0.0113563 (rank : 5) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
CPSF6_HUMAN
|
||||||
| NC score | 0.116210 (rank : 5) | θ value | 0.0563607 (rank : 6) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CPSF6_MOUSE
|
||||||
| NC score | 0.115578 (rank : 6) | θ value | 0.0563607 (rank : 7) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.114719 (rank : 7) | θ value | 0.00134147 (rank : 3) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
CPSF7_HUMAN
|
||||||
| NC score | 0.092563 (rank : 8) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N684, Q7Z3H9, Q9H025, Q9H9V1 | Gene names | CPSF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7 (Cleavage and polyadenylation specificity factor 59 kDa subunit) (CPSF 59 kDa subunit) (Pre-mRNA cleavage factor Im 59 kDa subunit). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.092258 (rank : 9) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
CPSF7_MOUSE
|
||||||
| NC score | 0.092240 (rank : 10) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BTV2, Q3TNF1, Q8BKE7, Q8CFS8 | Gene names | Cpsf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7. | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.080069 (rank : 11) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.076347 (rank : 12) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.070084 (rank : 13) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.067585 (rank : 14) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.066053 (rank : 15) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
FBX7_HUMAN
|
||||||
| NC score | 0.065763 (rank : 16) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y3I1, Q5TGC4, Q96HM6, Q9UF21, Q9UKT2 | Gene names | FBXO7, FBX7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 7. | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.063533 (rank : 17) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.062796 (rank : 18) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.060331 (rank : 19) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.060257 (rank : 20) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
RSMN_HUMAN
|
||||||
| NC score | 0.060044 (rank : 21) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P63162, P14648, P17135 | Gene names | SNRPN, HCERN3, SMN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
RSMN_MOUSE
|
||||||
| NC score | 0.060044 (rank : 22) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P63163, P14648, P17135 | Gene names | Snrpn, Smn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.058851 (rank : 23) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.058349 (rank : 24) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.058094 (rank : 25) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.054222 (rank : 26) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.052887 (rank : 27) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
TPO_MOUSE
|
||||||
| NC score | 0.052565 (rank : 28) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.052402 (rank : 29) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.052354 (rank : 30) | θ value | 0.21417 (rank : 9) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.051604 (rank : 31) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.051550 (rank : 32) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.051547 (rank : 33) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.051227 (rank : 34) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
RSMB_HUMAN
|
||||||
| NC score | 0.050677 (rank : 35) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P14678, Q15490, Q9UIS5 | Gene names | SNRPB, COD, SNRPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Small nuclear ribonucleoprotein-associated proteins B and B' (snRNP-B) (Sm protein B/B') (Sm-B/Sm-B') (SmB/SmB'). | |||||
|
RSMB_MOUSE
|
||||||
| NC score | 0.050480 (rank : 36) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P27048, Q3UJT1 | Gene names | Snrpb | |||
|
Domain Architecture |

|
|||||
| Description | Small nuclear ribonucleoprotein-associated protein B (snRNP-B) (Sm protein B) (Sm-B) (SmB). | |||||
|
PP1RA_HUMAN
|
||||||
| NC score | 0.048453 (rank : 37) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96QC0, O00405 | Gene names | PPP1R10, CAT53, FB19, PNUTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (Phosphatase 1 nuclear targeting subunit) (MHC class I region proline- rich protein CAT53) (FB19 protein) (PP1-binding protein of 114 kDa) (p99). | |||||
|
PB1_HUMAN
|
||||||
| NC score | 0.047178 (rank : 38) | θ value | 0.365318 (rank : 10) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.041923 (rank : 39) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.041770 (rank : 40) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
WASF3_MOUSE
|
||||||
| NC score | 0.041243 (rank : 41) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VHI6 | Gene names | Wasf3, Wave3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3). | |||||
|
WASF4_HUMAN
|
||||||
| NC score | 0.040892 (rank : 42) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.039278 (rank : 43) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.038424 (rank : 44) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WASF2_HUMAN
|
||||||
| NC score | 0.037803 (rank : 45) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
DAZP1_HUMAN
|
||||||
| NC score | 0.037306 (rank : 46) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96EP5, Q96MJ3, Q9NRR9 | Gene names | DAZAP1 | |||
|
Domain Architecture |

|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
SF3B4_HUMAN
|
||||||
| NC score | 0.037271 (rank : 47) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
DAZP1_MOUSE
|
||||||
| NC score | 0.037222 (rank : 48) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JII5 | Gene names | Dazap1 | |||
|
Domain Architecture |

|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.037055 (rank : 49) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
PP1RA_MOUSE
|
||||||
| NC score | 0.036635 (rank : 50) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80W00, Q811B6, Q8C6T7, Q8K2U8 | Gene names | Ppp1r10, Cat53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (MHC class I region proline-rich protein CAT53). | |||||
|
DYN1_MOUSE
|
||||||
| NC score | 0.036521 (rank : 51) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P39053, Q61358, Q61359, Q61360, Q8JZZ4, Q9CSY7, Q9QXX1 | Gene names | Dnm1, Dnm | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
DYN1_HUMAN
|
||||||
| NC score | 0.036494 (rank : 52) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q05193 | Gene names | DNM1, DNM | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-1 (EC 3.6.5.5). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.036302 (rank : 53) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.036296 (rank : 54) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.034650 (rank : 55) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.034520 (rank : 56) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
EPN1_MOUSE
|
||||||
| NC score | 0.033754 (rank : 57) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.032129 (rank : 58) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.030568 (rank : 59) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
NFIC_HUMAN
|
||||||
| NC score | 0.026505 (rank : 60) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08651, P08652, Q14932, Q9UPJ3, Q9UPJ9, Q9UPK0, Q9UPK1 | Gene names | NFIC, NFI | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 C-type (Nuclear factor 1/C) (NF1-C) (NFI-C) (NF-I/C) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
RBM16_HUMAN
|
||||||
| NC score | 0.025843 (rank : 61) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
TENS1_HUMAN
|
||||||
| NC score | 0.025566 (rank : 62) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
G3BP_MOUSE
|
||||||
| NC score | 0.024173 (rank : 63) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97855 | Gene names | G3bp, G3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 1 (EC 3.6.1.-) (ATP- dependent DNA helicase VIII) (GAP SH3-domain-binding protein 1) (G3BP- 1) (HDH-VIII). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.023993 (rank : 64) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.023432 (rank : 65) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
SC24D_HUMAN
|
||||||
| NC score | 0.023102 (rank : 66) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94855 | Gene names | SEC24D, KIAA0755 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24D (SEC24-related protein D). | |||||
|
RXRB_HUMAN
|
||||||
| NC score | 0.022429 (rank : 67) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P28702, P28703 | Gene names | RXRB, NR2B2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-beta (Retinoid X receptor beta). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.022395 (rank : 68) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
MCM3A_MOUSE
|
||||||
| NC score | 0.022216 (rank : 69) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WUU9 | Gene names | Mcm3ap, Ganp, Map80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
FUBP1_HUMAN
|
||||||
| NC score | 0.021509 (rank : 70) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96AE4, Q12828 | Gene names | FUBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP) (DNA helicase V) (HDH V). | |||||
|
FUBP1_MOUSE
|
||||||
| NC score | 0.021421 (rank : 71) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91WJ8, Q8C0Y8 | Gene names | Fubp1, D3Ertd330e | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP). | |||||
|
DAAM1_MOUSE
|
||||||
| NC score | 0.021145 (rank : 72) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BPM0, Q80Y68, Q9CQQ2 | Gene names | Daam1 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.021028 (rank : 73) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.020862 (rank : 74) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
CO2A1_MOUSE
|
||||||
| NC score | 0.020560 (rank : 75) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P28481 | Gene names | Col2a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
LAP4_HUMAN
|
||||||
| NC score | 0.019780 (rank : 76) | θ value | 0.125558 (rank : 8) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
FMN1B_MOUSE
|
||||||
| NC score | 0.019320 (rank : 77) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05859 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoform IV (Limb deformity protein). | |||||
|
FMN1A_MOUSE
|
||||||
| NC score | 0.018800 (rank : 78) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q05860 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoforms I/II/III (Limb deformity protein). | |||||
|
TNFL6_MOUSE
|
||||||
| NC score | 0.017712 (rank : 79) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41047, Q61217, Q9R1F2 | Gene names | Faslg, Apt1lg1, Fasl, gld, Tnfsf6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor ligand superfamily member 6 (Fas antigen ligand) (Fas ligand) (CD178 antigen) (CD95L protein) [Contains: Tumor necrosis factor ligand superfamily member 6, membrane form; Tumor necrosis factor ligand superfamily member 6, soluble form]. | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.017239 (rank : 80) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
SF01_HUMAN
|
||||||
| NC score | 0.017130 (rank : 81) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.016193 (rank : 82) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.016074 (rank : 83) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
3BP1_HUMAN
|
||||||
| NC score | 0.015741 (rank : 84) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
BAI1_HUMAN
|
||||||
| NC score | 0.015621 (rank : 85) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
CHGUT_HUMAN
|
||||||
| NC score | 0.015443 (rank : 86) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P2E5, Q6P2I4, Q6UXD2 | Gene names | CSGLCAT, KIAA1402 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate glucuronyltransferase (EC 2.4.1.226) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (Chondroitin glucuronyltransferase II) (CSGlcA-T). | |||||
|
BAI1_MOUSE
|
||||||
| NC score | 0.015104 (rank : 87) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3UHD1, Q3UH36, Q8CGM0 | Gene names | Bai1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.013988 (rank : 88) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
RXRB_MOUSE
|
||||||
| NC score | 0.013983 (rank : 89) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28704, P33243 | Gene names | Rxrb, Nr2b2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-beta (Retinoid X receptor beta) (MHC class I regulatory element-binding protein H-2RIIBP). | |||||
|
CO3A1_MOUSE
|
||||||
| NC score | 0.013978 (rank : 90) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
C1QRF_MOUSE
|
||||||
| NC score | 0.013621 (rank : 91) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88992 | Gene names | C1ql1, C1qrf, Crf | |||
|
Domain Architecture |

|
|||||
| Description | C1q-related factor precursor (Complement component 1 Q subcomponent- like 1). | |||||
|
IKBE_MOUSE
|
||||||
| NC score | 0.013388 (rank : 92) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54910, Q3U686, Q9CZZ9, Q9D7U3 | Gene names | Nfkbie, Ikbe | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKB-E). | |||||
|
ESX1L_HUMAN
|
||||||
| NC score | 0.013168 (rank : 93) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N693, Q7Z6K7 | Gene names | ESX1L, ESX1R | |||
|
Domain Architecture |

|
|||||
| Description | Extraembryonic, spermatogenesis, homeobox 1-like protein. | |||||
|
COIA1_MOUSE
|
||||||
| NC score | 0.012347 (rank : 94) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
AATK_HUMAN
|
||||||
| NC score | 0.010959 (rank : 95) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
BT2A3_HUMAN
|
||||||
| NC score | 0.010805 (rank : 96) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96KV6 | Gene names | BTN2A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Butyrophilin subfamily 2 member A3 precursor. | |||||
|
DOCK8_HUMAN
|
||||||
| NC score | 0.009766 (rank : 97) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NF50, Q3MV16, Q5JPJ1, Q8TEP1, Q8WUY2, Q9BYJ5, Q9H1Q2, Q9H1Q3, Q9H308, Q9H7P2 | Gene names | DOCK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 8. | |||||
|
MK07_HUMAN
|
||||||
| NC score | -0.000322 (rank : 98) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||