Please be patient as the page loads
|
B4GN4_HUMAN
|
||||||
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
B4GN4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 131 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GN4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.962859 (rank : 2) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GN3_MOUSE
|
||||||
| θ value | 6.86991e-125 (rank : 3) | NC score | 0.903593 (rank : 4) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6L8S8 | Gene names | B4galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
B4GN3_HUMAN
|
||||||
| θ value | 4.31301e-119 (rank : 4) | NC score | 0.906427 (rank : 3) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6L9W6, Q6ZNC1, Q8N7T6 | Gene names | B4GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
CGAT1_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 5) | NC score | 0.576560 (rank : 5) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TDX6, Q6P9G6, Q8IUF9, Q9NSQ7, Q9NUM9 | Gene names | GALNACT1, CHGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT1_MOUSE
|
||||||
| θ value | 8.95645e-16 (rank : 6) | NC score | 0.574357 (rank : 6) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BJQ9, Q3UZX6, Q8BWV9, Q8BZU7, Q8C195, Q8R0M6 | Gene names | Galnact1, Chgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT2_HUMAN
|
||||||
| θ value | 8.38298e-14 (rank : 7) | NC score | 0.560313 (rank : 7) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N6G5, Q6MZJ5, Q6MZP6, Q8TCH4, Q9P1I6 | Gene names | GALNACT2, CHGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CGAT2_MOUSE
|
||||||
| θ value | 3.18553e-13 (rank : 8) | NC score | 0.553669 (rank : 8) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C1F4, Q6PAP6, Q8R5A2, Q9D2M1 | Gene names | Galnact2, Chgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CHSS1_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 9) | NC score | 0.492594 (rank : 9) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86X52, Q6UX38, Q7LFU5, Q9Y2J5 | Gene names | CHSY1, CHSY, CSS1, KIAA0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase 1) (Chondroitin sulfate synthase 1) (N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS1_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 10) | NC score | 0.489091 (rank : 10) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS3_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 11) | NC score | 0.471310 (rank : 11) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 12) | NC score | 0.056383 (rank : 69) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 13) | NC score | 0.030046 (rank : 135) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
MAGD4_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 14) | NC score | 0.034896 (rank : 125) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 15) | NC score | 0.044135 (rank : 110) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CHSS2_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 16) | NC score | 0.228599 (rank : 13) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6IQX7 | Gene names | Css2, D1Bwg1363e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase). | |||||
|
SEM6C_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 17) | NC score | 0.025981 (rank : 140) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 18) | NC score | 0.042185 (rank : 114) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
CHSS2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 19) | NC score | 0.230277 (rank : 12) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IZ52, Q6UXD6, Q7L4G1, Q9H0F8, Q9H618 | Gene names | CSS2, CHPF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase) (Chondroitin-polymerizing factor). | |||||
|
SP5_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 20) | NC score | 0.010975 (rank : 168) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
TRM1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 21) | NC score | 0.055681 (rank : 79) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TX08, Q3TBY4 | Gene names | Trmt1, D8Ertd812e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N(2),N(2)-dimethylguanosine tRNA methyltransferase (EC 2.1.1.32) (tRNA(guanine-26,N(2)-N(2)) methyltransferase) (tRNA 2,2- dimethylguanosine-26 methyltransferase) (tRNA(m(2,2)G26)dimethyltransferase). | |||||
|
B4GT6_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 22) | NC score | 0.102921 (rank : 17) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UBX8, O60514 | Gene names | B4GALT6 | |||
|
Domain Architecture |
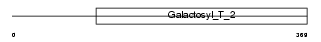
|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT6_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 23) | NC score | 0.102437 (rank : 18) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WVK5 | Gene names | B4galt6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 24) | NC score | 0.040212 (rank : 118) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
B4GT5_HUMAN
|
||||||
| θ value | 0.125558 (rank : 25) | NC score | 0.097105 (rank : 21) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43286, Q2M394, Q9UJQ8 | Gene names | B4GALT5 | |||
|
Domain Architecture |
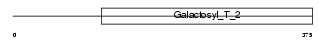
|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT5_MOUSE
|
||||||
| θ value | 0.125558 (rank : 26) | NC score | 0.097489 (rank : 20) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JMK0 | Gene names | B4galt5, Bgt-5 | |||
|
Domain Architecture |
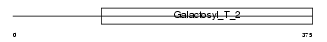
|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
GAK12_HUMAN
|
||||||
| θ value | 0.125558 (rank : 27) | NC score | 0.067569 (rank : 39) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GALT3_MOUSE
|
||||||
| θ value | 0.125558 (rank : 28) | NC score | 0.073673 (rank : 31) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70419, Q80V55 | Gene names | Galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 3) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 3) (Polypeptide GalNAc transferase 3) (GalNAc-T3) (pp-GaNTase 3). | |||||
|
RUSC1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 29) | NC score | 0.048042 (rank : 107) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BG26, Q6PHT9 | Gene names | Rusc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1. | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.125558 (rank : 30) | NC score | 0.038128 (rank : 122) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
B4GT2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 31) | NC score | 0.121230 (rank : 15) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60909, O60511, Q5T4X5, Q5T4Y5, Q9BUP6, Q9NSY7 | Gene names | B4GALT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
GAK11_HUMAN
|
||||||
| θ value | 0.163984 (rank : 32) | NC score | 0.066159 (rank : 43) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 33) | NC score | 0.066184 (rank : 42) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 34) | NC score | 0.066247 (rank : 41) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
SP5_HUMAN
|
||||||
| θ value | 0.163984 (rank : 35) | NC score | 0.009372 (rank : 174) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
B4GT2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 36) | NC score | 0.119972 (rank : 16) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z2Y2, Q3TMP2 | Gene names | B4galt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
DMRTD_HUMAN
|
||||||
| θ value | 0.21417 (rank : 37) | NC score | 0.043910 (rank : 111) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IXT2, Q8N6Q2, Q96M39, Q96SD4 | Gene names | DMRTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2. | |||||
|
GALT3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 38) | NC score | 0.071857 (rank : 33) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14435, Q7Z476 | Gene names | GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 3) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 3) (Polypeptide GalNAc transferase 3) (GalNAc-T3) (pp-GaNTase 3). | |||||
|
PP16A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 39) | NC score | 0.016788 (rank : 155) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96I34 | Gene names | PPP1R16A, MYPT3 | |||
|
Domain Architecture |
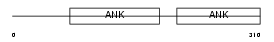
|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase- targeting subunit 3). | |||||
|
GALT6_HUMAN
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.070645 (rank : 35) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8NCL4, Q8IYH4, Q9H6G2, Q9UIV5 | Gene names | GALNT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
XKR7_MOUSE
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.026663 (rank : 139) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5GH64 | Gene names | Xkr7, Xrg7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
ULK1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 42) | NC score | 0.004089 (rank : 181) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
GA2L2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 43) | NC score | 0.040776 (rank : 116) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
GAK5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.064390 (rank : 51) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 45) | NC score | 0.064458 (rank : 50) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
ADA12_MOUSE
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.010761 (rank : 169) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.034674 (rank : 128) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
GAK10_HUMAN
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.064662 (rank : 49) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GALT6_MOUSE
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.065542 (rank : 44) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8C7U7, Q3TWF0, Q8CED2, Q9QZ16 | Gene names | Galnt6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.022185 (rank : 147) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.018374 (rank : 151) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
ZAR1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.042784 (rank : 112) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86SH2 | Gene names | ZAR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.030319 (rank : 133) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MINT_MOUSE
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.034728 (rank : 127) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MK15_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.003749 (rank : 182) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TD08, Q2TCF9, Q8N362 | Gene names | MAPK15, ERK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 8). | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.013122 (rank : 162) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.042046 (rank : 115) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.017361 (rank : 153) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.051377 (rank : 101) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.031771 (rank : 132) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.040606 (rank : 117) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.011277 (rank : 167) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
T240L_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.033964 (rank : 130) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q71F56, Q68DN4, Q9H8C0, Q9NSY9, Q9UFD8, Q9UPX5 | Gene names | THRAP2, KIAA1025, PROSIT240, TRAP240L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
TXLNA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.012599 (rank : 163) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
3BP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.045224 (rank : 109) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
GLT14_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.055998 (rank : 74) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96FL9, Q8IVI4, Q9BRH1, Q9H827, Q9H9J8 | Gene names | GALNT14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 14 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 14) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 14) (Polypeptide GalNAc transferase 14) (GalNAc-T14) (pp-GaNTase 14). | |||||
|
GLT14_MOUSE
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.055880 (rank : 76) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BVG5, Q60GT2, Q8BTI6, Q9DCJ2 | Gene names | Galnt14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 14 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 14) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 14) (Polypeptide GalNAc transferase 14) (GalNAc-T14) (pp-GaNTase 14). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.056042 (rank : 73) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PER1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.038636 (rank : 120) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.035463 (rank : 124) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
ZO1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.015547 (rank : 156) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
CELR3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.002736 (rank : 184) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
DOCK4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.023097 (rank : 145) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.023617 (rank : 144) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
GAK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.062564 (rank : 54) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GALT5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.055237 (rank : 80) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8C102 | Gene names | Galnt5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.027878 (rank : 137) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.038099 (rank : 123) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.018562 (rank : 150) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.025043 (rank : 141) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
TTC16_HUMAN
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.020348 (rank : 148) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
DCC_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.007165 (rank : 179) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
ELOA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.020013 (rank : 149) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IYF1, Q9P2V9 | Gene names | TCEB3B, TCEB3L | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A2 (Elongin-A2) (EloA2) (Transcription elongation factor B polypeptide 3B). | |||||
|
FLNC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.009085 (rank : 176) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14315, O95303, Q07985, Q9NS12, Q9NYE5, Q9UMR8, Q9Y503 | Gene names | FLNC, ABPL, FLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-C (Gamma-filamin) (Filamin-2) (Protein FLNc) (Actin-binding- like protein) (ABP-L) (ABP-280-like protein). | |||||
|
GALT4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.056785 (rank : 63) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N4A0, O00208 | Gene names | GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 4) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 4) (Polypeptide GalNAc transferase 4) (GalNAc-T4) (pp-GaNTase 4). | |||||
|
GALT4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.056770 (rank : 64) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08832, Q3U681 | Gene names | Galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 4) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 4) (Polypeptide GalNAc transferase 4) (GalNAc-T4) (pp-GaNTase 4). | |||||
|
GALT5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.055014 (rank : 82) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7Z7M9, Q9UGK7, Q9UHL6 | Gene names | GALNT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
GLT12_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.057267 (rank : 61) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BGT9 | Gene names | Galnt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 12 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 12) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 12) (Polypeptide GalNAc transferase 12) (GalNAc-T12) (pp-GaNTase 12). | |||||
|
GLTL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.055762 (rank : 78) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N428, Q9ULT9 | Gene names | GALNTL1, KIAA1130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 1 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 1) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 1) (Polypeptide GalNAc transferase-like protein 1) (GalNAc-T-like protein 1) (pp-GaNTase-like protein 1). | |||||
|
GLTL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.055800 (rank : 77) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JJ61 | Gene names | Galntl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 1 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 1) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 1) (Polypeptide GalNAc transferase-like protein 1) (GalNAc-T-like protein 1) (pp-GaNTase-like protein 1). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.017269 (rank : 154) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.034062 (rank : 129) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PAK4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.003727 (rank : 183) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1098 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BTW9, Q6ZPX0, Q80Z97, Q9CS71 | Gene names | Pak4, Kiaa1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||
|
CAN9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.006306 (rank : 180) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14815, Q9NS74 | Gene names | CAPN9, NCL4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4) (CG36 protein). | |||||
|
HAIR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.014526 (rank : 159) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
LAT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.024676 (rank : 143) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54957 | Gene names | Lat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Linker for activation of T-cells family member 1 (36 kDa phospho- tyrosine adapter protein) (pp36) (p36-38). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.027603 (rank : 138) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MYO15_MOUSE
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.012185 (rank : 166) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
AMOT_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.028085 (rank : 136) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
CAR10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.009268 (rank : 175) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P58660 | Gene names | Card10, Bimp1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 10 (Bcl10-interacting MAGUK protein 1) (Bimp1). | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.008287 (rank : 177) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
GLT13_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.053426 (rank : 91) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IUC8, Q6ZWG1, Q96PX0, Q9UIE5 | Gene names | GALNT13, KIAA1918 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 13 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 13) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 13) (Polypeptide GalNAc transferase 13) (GalNAc-T13) (pp-GaNTase 13). | |||||
|
GLT13_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.053440 (rank : 90) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CF93, Q8BLE4, Q8BYT3 | Gene names | Galnt13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 13 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 13) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 13) (Polypeptide GalNAc transferase 13) (GalNAc-T13) (pp-GaNTase 13). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.077775 (rank : 28) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.015126 (rank : 157) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.010538 (rank : 172) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.040060 (rank : 119) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
TCAL3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.022863 (rank : 146) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
CD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.033351 (rank : 131) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
CS007_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.034738 (rank : 126) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
GASP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.012487 (rank : 165) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96D09, Q8NAB4 | Gene names | GPRASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.012512 (rank : 164) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.042303 (rank : 113) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MUS81_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.013761 (rank : 160) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96NY9, Q9H7D9 | Gene names | MUS81 | |||
|
Domain Architecture |

|
|||||
| Description | Crossover junction endonuclease MUS81 (EC 3.1.22.-). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.024758 (rank : 142) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SC6A5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.008089 (rank : 178) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q761V0, Q8CFM5, Q91ZQ2 | Gene names | Slc6a5, Glyt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
B4GT3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.064716 (rank : 48) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60512, O60910, Q9BPZ4, Q9H8T2 | Gene names | B4GALT3 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.065185 (rank : 46) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91YY2, Q9QY13 | Gene names | B4galt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.030156 (rank : 134) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CBL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.009947 (rank : 173) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P22681 | Gene names | CBL, CBL2, RNF55 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene) (RING finger protein 55). | |||||
|
CK024_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.047722 (rank : 108) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CO039_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.038131 (rank : 121) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
DBND2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.013313 (rank : 161) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQY9, Q9BQZ0, Q9BVL1, Q9H1F6, Q9NWZ0, Q9NY07, Q9NZ31 | Gene names | DBNDD2, C20orf35 | |||
|
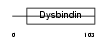
Domain Architecture |
|
|||||
| Description | Dysbindin domain-containing protein 2 (HSMNP1). | |||||
|
GLT12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.054245 (rank : 85) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IXK2, Q5TCF7, Q8NG54, Q96CT9, Q9H771 | Gene names | GALNT12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 12 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 12) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 12) (Polypeptide GalNAc transferase 12) (GalNAc-T12) (pp-GaNTase 12). | |||||
|
IF32_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.010658 (rank : 170) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13347 | Gene names | EIF3S2, TRIP1 | |||
|
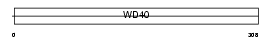
Domain Architecture |
|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 2 (eIF-3 beta) (eIF3 p36) (eIF3i) (TGF-beta receptor-interacting protein 1) (TRIP-1). | |||||
|
IF32_MOUSE
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.010656 (rank : 171) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZD9 | Gene names | Eif3s2, Trip1 | |||
|
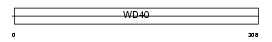
Domain Architecture |
|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 2 (eIF-3 beta) (eIF3 p36) (eIF3i) (TGF-beta receptor-interacting protein 1) (TRIP-1). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.018358 (rank : 152) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
MK04_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.001085 (rank : 186) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6P5G0 | Gene names | Mapk4, Erk4, Prkm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 4 (EC 2.7.11.24) (Extracellular signal-regulated kinase 4) (ERK-4). | |||||
|
PAK7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.001264 (rank : 185) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C015, Q3TQJ7, Q6RWS7 | Gene names | Pak7, Pak5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PAK 7 (EC 2.7.11.1) (p21-activated kinase 7) (PAK-7) (p21-activated kinase 5). | |||||
|
PALM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.014803 (rank : 158) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
ZFP13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | -0.001630 (rank : 187) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10754, Q80WS0, Q8CDD0 | Gene names | Zfp13, Krox-8, Zfp-13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 13 (Zfp-13) (Krox-8 protein). | |||||
|
B3GLT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.052678 (rank : 95) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6Y288, Q5W0H2, Q6NUI3 | Gene names | B3GALTL, B3GTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
B3GLT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.056904 (rank : 62) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHT6, Q0PCR7, Q149S5 | Gene names | B3galtl, Gm1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
B4GT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.070994 (rank : 34) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.077835 (rank : 27) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15535 | Gene names | B4galt1, Ggtb, Ggtb2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.070038 (rank : 36) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60513, Q68D68, Q9BSW3, Q9C078 | Gene names | B4GALT4 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.071868 (rank : 32) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JJ04, Q8BR54, Q9QY12 | Gene names | B4galt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.051462 (rank : 100) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UBV7, Q9UHN2 | Gene names | B4GALT7, XGALT1 | |||
|
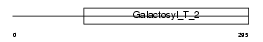
Domain Architecture |
|
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
B4GT7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.052265 (rank : 97) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R087, Q7TNU3 | Gene names | B4galt7, Xgalt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.054343 (rank : 84) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.051961 (rank : 98) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CHGUT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.190445 (rank : 14) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2E5, Q6P2I4, Q6UXD2 | Gene names | CSGLCAT, KIAA1402 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate glucuronyltransferase (EC 2.4.1.226) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (Chondroitin glucuronyltransferase II) (CSGlcA-T). | |||||
|
CN032_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.060462 (rank : 56) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.056545 (rank : 68) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CPSF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.050672 (rank : 105) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CPSF6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.051835 (rank : 99) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.053743 (rank : 87) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
GAK15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.052507 (rank : 96) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62688 | Gene names | ERVK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q21.2 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(I) Gag protein). | |||||
|
GAK17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.052821 (rank : 93) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62689 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein]. | |||||
|
GAK18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.056589 (rank : 67) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK19_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.050674 (rank : 104) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P62691 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_4p16.1 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GAK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.057798 (rank : 60) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.054121 (rank : 86) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96PI4, Q9QC08 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K18 Gag protein) (HERV-K110 Gag protein) (HERV-K(C1a) Gag protein) [Contains: Matrix protein]. | |||||
|
GAK8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.056598 (rank : 66) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HDB9 | Gene names | ERVK5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q12.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(II) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.052742 (rank : 94) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UKH8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K104 Gag protein) [Contains: Matrix protein]. | |||||
|
GGN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.063098 (rank : 52) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GGN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.055231 (rank : 81) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.056274 (rank : 70) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.056244 (rank : 71) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.067283 (rank : 40) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.082088 (rank : 26) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.086762 (rank : 24) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.059287 (rank : 58) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.084320 (rank : 25) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.091079 (rank : 23) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.068400 (rank : 38) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.053341 (rank : 92) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.051374 (rank : 102) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.062781 (rank : 53) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.060491 (rank : 55) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.050392 (rank : 106) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SFR15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.053527 (rank : 89) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
SMR3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.056739 (rank : 65) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.098763 (rank : 19) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.092903 (rank : 22) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.068982 (rank : 37) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
WASF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.051022 (rank : 103) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.077699 (rank : 29) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.076436 (rank : 30) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WASL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.053688 (rank : 88) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WASL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.065055 (rank : 47) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.056090 (rank : 72) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
WASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.055949 (rank : 75) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.059468 (rank : 57) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.058729 (rank : 59) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.065466 (rank : 45) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
WIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.054616 (rank : 83) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
B4GN4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 131 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GN4_MOUSE
|
||||||
| NC score | 0.962859 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GN3_HUMAN
|
||||||
| NC score | 0.906427 (rank : 3) | θ value | 4.31301e-119 (rank : 4) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6L9W6, Q6ZNC1, Q8N7T6 | Gene names | B4GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
B4GN3_MOUSE
|
||||||
| NC score | 0.903593 (rank : 4) | θ value | 6.86991e-125 (rank : 3) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6L8S8 | Gene names | B4galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
CGAT1_HUMAN
|
||||||
| NC score | 0.576560 (rank : 5) | θ value | 1.80466e-16 (rank : 5) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TDX6, Q6P9G6, Q8IUF9, Q9NSQ7, Q9NUM9 | Gene names | GALNACT1, CHGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT1_MOUSE
|
||||||
| NC score | 0.574357 (rank : 6) | θ value | 8.95645e-16 (rank : 6) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BJQ9, Q3UZX6, Q8BWV9, Q8BZU7, Q8C195, Q8R0M6 | Gene names | Galnact1, Chgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT2_HUMAN
|
||||||
| NC score | 0.560313 (rank : 7) | θ value | 8.38298e-14 (rank : 7) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N6G5, Q6MZJ5, Q6MZP6, Q8TCH4, Q9P1I6 | Gene names | GALNACT2, CHGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CGAT2_MOUSE
|
||||||
| NC score | 0.553669 (rank : 8) | θ value | 3.18553e-13 (rank : 8) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C1F4, Q6PAP6, Q8R5A2, Q9D2M1 | Gene names | Galnact2, Chgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CHSS1_HUMAN
|
||||||
| NC score | 0.492594 (rank : 9) | θ value | 7.09661e-13 (rank : 9) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86X52, Q6UX38, Q7LFU5, Q9Y2J5 | Gene names | CHSY1, CHSY, CSS1, KIAA0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase 1) (Chondroitin sulfate synthase 1) (N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS1_MOUSE
|
||||||
| NC score | 0.489091 (rank : 10) | θ value | 2.69671e-12 (rank : 10) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS3_HUMAN
|
||||||
| NC score | 0.471310 (rank : 11) | θ value | 2.79066e-09 (rank : 11) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
CHSS2_HUMAN
|
||||||
| NC score | 0.230277 (rank : 12) | θ value | 0.0431538 (rank : 19) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IZ52, Q6UXD6, Q7L4G1, Q9H0F8, Q9H618 | Gene names | CSS2, CHPF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase) (Chondroitin-polymerizing factor). | |||||
|
CHSS2_MOUSE
|
||||||
| NC score | 0.228599 (rank : 13) | θ value | 0.0148317 (rank : 16) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6IQX7 | Gene names | Css2, D1Bwg1363e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase). | |||||
|
CHGUT_HUMAN
|
||||||
| NC score | 0.190445 (rank : 14) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2E5, Q6P2I4, Q6UXD2 | Gene names | CSGLCAT, KIAA1402 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate glucuronyltransferase (EC 2.4.1.226) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (Chondroitin glucuronyltransferase II) (CSGlcA-T). | |||||
|
B4GT2_HUMAN
|
||||||
| NC score | 0.121230 (rank : 15) | θ value | 0.163984 (rank : 31) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60909, O60511, Q5T4X5, Q5T4Y5, Q9BUP6, Q9NSY7 | Gene names | B4GALT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT2_MOUSE
|
||||||
| NC score | 0.119972 (rank : 16) | θ value | 0.21417 (rank : 36) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z2Y2, Q3TMP2 | Gene names | B4galt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT6_HUMAN
|
||||||
| NC score | 0.102921 (rank : 17) | θ value | 0.0736092 (rank : 22) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UBX8, O60514 | Gene names | B4GALT6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT6_MOUSE
|
||||||
| NC score | 0.102437 (rank : 18) | θ value | 0.0961366 (rank : 23) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WVK5 | Gene names | B4galt6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.098763 (rank : 19) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
B4GT5_MOUSE
|
||||||
| NC score | 0.097489 (rank : 20) | θ value | 0.125558 (rank : 26) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JMK0 | Gene names | B4galt5, Bgt-5 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT5_HUMAN
|
||||||
| NC score | 0.097105 (rank : 21) | θ value | 0.125558 (rank : 25) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43286, Q2M394, Q9UJQ8 | Gene names | B4GALT5 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.092903 (rank : 22) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.091079 (rank : 23) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.086762 (rank : 24) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.084320 (rank : 25) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.082088 (rank : 26) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
B4GT1_MOUSE
|
||||||
| NC score | 0.077835 (rank : 27) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15535 | Gene names | B4galt1, Ggtb, Ggtb2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.077775 (rank : 28) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.077699 (rank : 29) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.076436 (rank : 30) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
GALT3_MOUSE
|
||||||
| NC score | 0.073673 (rank : 31) | θ value | 0.125558 (rank : 28) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70419, Q80V55 | Gene names | Galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 3) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 3) (Polypeptide GalNAc transferase 3) (GalNAc-T3) (pp-GaNTase 3). | |||||
|
B4GT4_MOUSE
|
||||||
| NC score | 0.071868 (rank : 32) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JJ04, Q8BR54, Q9QY12 | Gene names | B4galt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
GALT3_HUMAN
|
||||||
| NC score | 0.071857 (rank : 33) | θ value | 0.21417 (rank : 38) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14435, Q7Z476 | Gene names | GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 3) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 3) (Polypeptide GalNAc transferase 3) (GalNAc-T3) (pp-GaNTase 3). | |||||
|
B4GT1_HUMAN
|
||||||
| NC score | 0.070994 (rank : 34) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
GALT6_HUMAN
|
||||||
| NC score | 0.070645 (rank : 35) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8NCL4, Q8IYH4, Q9H6G2, Q9UIV5 | Gene names | GALNT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
B4GT4_HUMAN
|
||||||
| NC score | 0.070038 (rank : 36) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60513, Q68D68, Q9BSW3, Q9C078 | Gene names | B4GALT4 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.068982 (rank : 37) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.068400 (rank : 38) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
GAK12_HUMAN
|
||||||
| NC score | 0.067569 (rank : 39) | θ value | 0.125558 (rank : 27) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.067283 (rank : 40) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.066247 (rank : 41) | θ value | 0.163984 (rank : 34) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.066184 (rank : 42) | θ value | 0.163984 (rank : 33) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| NC score | 0.066159 (rank : 43) | θ value | 0.163984 (rank : 32) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GALT6_MOUSE
|
||||||
| NC score | 0.065542 (rank : 44) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8C7U7, Q3TWF0, Q8CED2, Q9QZ16 | Gene names | Galnt6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.065466 (rank : 45) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
B4GT3_MOUSE
|
||||||
| NC score | 0.065185 (rank : 46) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91YY2, Q9QY13 | Gene names | B4galt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.065055 (rank : 47) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
B4GT3_HUMAN
|
||||||
| NC score | 0.064716 (rank : 48) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60512, O60910, Q9BPZ4, Q9H8T2 | Gene names | B4GALT3 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
GAK10_HUMAN
|
||||||
| NC score | 0.064662 (rank : 49) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| NC score | 0.064458 (rank : 50) | θ value | 0.47712 (rank : 45) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK5_HUMAN
|
||||||
| NC score | 0.064390 (rank : 51) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GGN_HUMAN
|
||||||
| NC score | 0.063098 (rank : 52) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.062781 (rank : 53) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
GAK1_HUMAN
|
||||||
| NC score | 0.062564 (rank : 54) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.060491 (rank : 55) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.060462 (rank : 56) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.059468 (rank : 57) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.059287 (rank : 58) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.058729 (rank : 59) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
GAK3_HUMAN
|
||||||
| NC score | 0.057798 (rank : 60) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GLT12_MOUSE
|
||||||
| NC score | 0.057267 (rank : 61) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BGT9 | Gene names | Galnt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 12 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 12) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 12) (Polypeptide GalNAc transferase 12) (GalNAc-T12) (pp-GaNTase 12). | |||||
|
B3GLT_MOUSE
|
||||||
| NC score | 0.056904 (rank : 62) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHT6, Q0PCR7, Q149S5 | Gene names | B3galtl, Gm1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
GALT4_HUMAN
|
||||||
| NC score | 0.056785 (rank : 63) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N4A0, O00208 | Gene names | GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 4) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 4) (Polypeptide GalNAc transferase 4) (GalNAc-T4) (pp-GaNTase 4). | |||||
|
GALT4_MOUSE
|
||||||
| NC score | 0.056770 (rank : 64) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08832, Q3U681 | Gene names | Galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 4) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 4) (Polypeptide GalNAc transferase 4) (GalNAc-T4) (pp-GaNTase 4). | |||||
|
SMR3A_HUMAN
|
||||||
| NC score | 0.056739 (rank : 65) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
GAK8_HUMAN
|
||||||
| NC score | 0.056598 (rank : 66) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HDB9 | Gene names | ERVK5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q12.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(II) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK18_HUMAN
|
||||||
| NC score | 0.056589 (rank : 67) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.056545 (rank : 68) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.056383 (rank : 69) | θ value | 1.99992e-07 (rank : 12) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.056274 (rank : 70) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.056244 (rank : 71) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
WASP_HUMAN
|
||||||
| NC score | 0.056090 (rank : 72) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.056042 (rank : 73) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
GLT14_HUMAN
|
||||||
| NC score | 0.055998 (rank : 74) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96FL9, Q8IVI4, Q9BRH1, Q9H827, Q9H9J8 | Gene names | GALNT14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 14 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 14) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 14) (Polypeptide GalNAc transferase 14) (GalNAc-T14) (pp-GaNTase 14). | |||||
|
WASP_MOUSE
|
||||||
| NC score | 0.055949 (rank : 75) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
GLT14_MOUSE
|
||||||
| NC score | 0.055880 (rank : 76) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BVG5, Q60GT2, Q8BTI6, Q9DCJ2 | Gene names | Galnt14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 14 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 14) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 14) (Polypeptide GalNAc transferase 14) (GalNAc-T14) (pp-GaNTase 14). | |||||
|
GLTL1_MOUSE
|
||||||
| NC score | 0.055800 (rank : 77) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JJ61 | Gene names | Galntl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 1 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 1) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 1) (Polypeptide GalNAc transferase-like protein 1) (GalNAc-T-like protein 1) (pp-GaNTase-like protein 1). | |||||
|
GLTL1_HUMAN
|
||||||
| NC score | 0.055762 (rank : 78) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N428, Q9ULT9 | Gene names | GALNTL1, KIAA1130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 1 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 1) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 1) (Polypeptide GalNAc transferase-like protein 1) (GalNAc-T-like protein 1) (pp-GaNTase-like protein 1). | |||||
|
TRM1_MOUSE
|
||||||
| NC score | 0.055681 (rank : 79) | θ value | 0.0431538 (rank : 21) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TX08, Q3TBY4 | Gene names | Trmt1, D8Ertd812e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N(2),N(2)-dimethylguanosine tRNA methyltransferase (EC 2.1.1.32) (tRNA(guanine-26,N(2)-N(2)) methyltransferase) (tRNA 2,2- dimethylguanosine-26 methyltransferase) (tRNA(m(2,2)G26)dimethyltransferase). | |||||
|
GALT5_MOUSE
|
||||||
| NC score | 0.055237 (rank : 80) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8C102 | Gene names | Galnt5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.055231 (rank : 81) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GALT5_HUMAN
|
||||||
| NC score | 0.055014 (rank : 82) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7Z7M9, Q9UGK7, Q9UHL6 | Gene names | GALNT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.054616 (rank : 83) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.054343 (rank : 84) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
GLT12_HUMAN
|
||||||
| NC score | 0.054245 (rank : 85) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IXK2, Q5TCF7, Q8NG54, Q96CT9, Q9H771 | Gene names | GALNT12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 12 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 12) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 12) (Polypeptide GalNAc transferase 12) (GalNAc-T12) (pp-GaNTase 12). | |||||
|
GAK7_HUMAN
|
||||||
| NC score | 0.054121 (rank : 86) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96PI4, Q9QC08 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K18 Gag protein) (HERV-K110 Gag protein) (HERV-K(C1a) Gag protein) [Contains: Matrix protein]. | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.053743 (rank : 87) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.053688 (rank : 88) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
SFR15_HUMAN
|
||||||
| NC score | 0.053527 (rank : 89) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
GLT13_MOUSE
|
||||||
| NC score | 0.053440 (rank : 90) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CF93, Q8BLE4, Q8BYT3 | Gene names | Galnt13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 13 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 13) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 13) (Polypeptide GalNAc transferase 13) (GalNAc-T13) (pp-GaNTase 13). | |||||
|
GLT13_HUMAN
|
||||||
| NC score | 0.053426 (rank : 91) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IUC8, Q6ZWG1, Q96PX0, Q9UIE5 | Gene names | GALNT13, KIAA1918 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 13 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 13) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 13) (Polypeptide GalNAc transferase 13) (GalNAc-T13) (pp-GaNTase 13). | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.053341 (rank : 92) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
GAK17_HUMAN
|
||||||
| NC score | 0.052821 (rank : 93) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62689 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein]. | |||||
|
GAK9_HUMAN
|
||||||
| NC score | 0.052742 (rank : 94) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UKH8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K104 Gag protein) [Contains: Matrix protein]. | |||||
|
B3GLT_HUMAN
|
||||||
| NC score | 0.052678 (rank : 95) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6Y288, Q5W0H2, Q6NUI3 | Gene names | B3GALTL, B3GTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
GAK15_HUMAN
|
||||||
| NC score | 0.052507 (rank : 96) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P62688 | Gene names | ERVK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q21.2 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(I) Gag protein). | |||||
|
B4GT7_MOUSE
|
||||||
| NC score | 0.052265 (rank : 97) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R087, Q7TNU3 | Gene names | B4galt7, Xgalt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.051961 (rank : 98) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CPSF6_MOUSE
|
||||||
| NC score | 0.051835 (rank : 99) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
B4GT7_HUMAN
|
||||||
| NC score | 0.051462 (rank : 100) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UBV7, Q9UHN2 | Gene names | B4GALT7, XGALT1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.051377 (rank : 101) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.051374 (rank : 102) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
WASF2_HUMAN
|
||||||
| NC score | 0.051022 (rank : 103) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
GAK19_HUMAN
|
||||||
| NC score | 0.050674 (rank : 104) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P62691 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_4p16.1 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
CPSF6_HUMAN
|
||||||
| NC score | 0.050672 (rank : 105) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.050392 (rank : 106) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
RUSC1_MOUSE
|
||||||
| NC score | 0.048042 (rank : 107) | θ value | 0.125558 (rank : 29) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BG26, Q6PHT9 | Gene names | Rusc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1. | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.047722 (rank : 108) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
3BP2_HUMAN
|
||||||
| NC score | 0.045224 (rank : 109) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.044135 (rank : 110) | θ value | 0.00869519 (rank : 15) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
DMRTD_HUMAN
|
||||||
| NC score | 0.043910 (rank : 111) | θ value | 0.21417 (rank : 37) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IXT2, Q8N6Q2, Q96M39, Q96SD4 | Gene names | DMRTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2. | |||||
|
ZAR1_HUMAN
|
||||||
| NC score | 0.042784 (rank : 112) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86SH2 | Gene names | ZAR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.042303 (rank : 113) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.042185 (rank : 114) | θ value | 0.0330416 (rank : 18) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.042046 (rank : 115) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
GA2L2_MOUSE
|
||||||
| NC score | 0.040776 (rank : 116) | θ value | 0.47712 (rank : 43) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.040606 (rank : 117) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.040212 (rank : 118) | θ value | 0.0961366 (rank : 24) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.040060 (rank : 119) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
PER1_HUMAN
|
||||||
| NC score | 0.038636 (rank : 120) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.038131 (rank : 121) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.038128 (rank : 122) | θ value | 0.125558 (rank : 30) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.038099 (rank : 123) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
PER1_MOUSE
|
||||||
| NC score | 0.035463 (rank : 124) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
MAGD4_HUMAN
|
||||||
| NC score | 0.034896 (rank : 125) | θ value | 0.00509761 (rank : 14) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.034738 (rank : 126) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.034728 (rank : 127) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.034674 (rank : 128) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.034062 (rank : 129) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
T240L_HUMAN
|
||||||
| NC score | 0.033964 (rank : 130) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q71F56, Q68DN4, Q9H8C0, Q9NSY9, Q9UFD8, Q9UPX5 | Gene names | THRAP2, KIAA1025, PROSIT240, TRAP240L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
CD2_HUMAN
|
||||||
| NC score | 0.033351 (rank : 131) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.031771 (rank : 132) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.030319 (rank : 133) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.030156 (rank : 134) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.030046 (rank : 135) | θ value | 0.00228821 (rank : 13) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
AMOT_HUMAN
|
||||||
| NC score | 0.028085 (rank : 136) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.027878 (rank : 137) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.027603 (rank : 138) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
XKR7_MOUSE
|
||||||
| NC score | 0.026663 (rank : 139) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5GH64 | Gene names | Xkr7, Xrg7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
SEM6C_MOUSE
|
||||||
| NC score | 0.025981 (rank : 140) | θ value | 0.0193708 (rank : 17) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.025043 (rank : 141) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.024758 (rank : 142) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
LAT_MOUSE
|
||||||
| NC score | 0.024676 (rank : 143) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54957 | Gene names | Lat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Linker for activation of T-cells family member 1 (36 kDa phospho- tyrosine adapter protein) (pp36) (p36-38). | |||||
|
DOCK4_MOUSE
|
||||||
| NC score | 0.023617 (rank : 144) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK4_HUMAN
|
||||||
| NC score | 0.023097 (rank : 145) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
TCAL3_HUMAN
|
||||||
| NC score | 0.022863 (rank : 146) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.022185 (rank : 147) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
TTC16_HUMAN
|
||||||
| NC score | 0.020348 (rank : 148) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
ELOA2_HUMAN
|
||||||
| NC score | 0.020013 (rank : 149) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IYF1, Q9P2V9 | Gene names | TCEB3B, TCEB3L | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A2 (Elongin-A2) (EloA2) (Transcription elongation factor B polypeptide 3B). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.018562 (rank : 150) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.018374 (rank : 151) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.018358 (rank : 152) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.017361 (rank : 153) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.017269 (rank : 154) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
PP16A_HUMAN
|
||||||
| NC score | 0.016788 (rank : 155) | θ value | 0.21417 (rank : 39) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96I34 | Gene names | PPP1R16A, MYPT3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase- targeting subunit 3). | |||||
|
ZO1_HUMAN
|
||||||
| NC score | 0.015547 (rank : 156) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.015126 (rank : 157) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PALM_MOUSE
|
||||||
| NC score | 0.014803 (rank : 158) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z0P4, Q9Z0P3 | Gene names | Palm | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
HAIR_HUMAN
|
||||||
| NC score | 0.014526 (rank : 159) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
MUS81_HUMAN
|
||||||
| NC score | 0.013761 (rank : 160) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96NY9, Q9H7D9 | Gene names | MUS81 | |||
|
Domain Architecture |

|
|||||
| Description | Crossover junction endonuclease MUS81 (EC 3.1.22.-). | |||||
|
DBND2_HUMAN
|
||||||
| NC score | 0.013313 (rank : 161) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQY9, Q9BQZ0, Q9BVL1, Q9H1F6, Q9NWZ0, Q9NY07, Q9NZ31 | Gene names | DBNDD2, C20orf35 | |||
|
Domain Architecture |

|
|||||
| Description | Dysbindin domain-containing protein 2 (HSMNP1). | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.013122 (rank : 162) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
TXLNA_HUMAN
|
||||||
| NC score | 0.012599 (rank : 163) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P40222, Q66K62, Q86T54, Q86T85, Q86T86, Q86Y86, Q86YW3, Q8N2Y3 | Gene names | TXLNA, TXLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.012512 (rank : 164) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
GASP2_HUMAN
|
||||||
| NC score | 0.012487 (rank : 165) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96D09, Q8NAB4 | Gene names | GPRASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.012185 (rank : 166) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.011277 (rank : 167) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SP5_MOUSE
|
||||||
| NC score | 0.010975 (rank : 168) | θ value | 0.0431538 (rank : 20) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
ADA12_MOUSE
|
||||||
| NC score | 0.010761 (rank : 169) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
IF32_HUMAN
|
||||||
| NC score | 0.010658 (rank : 170) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13347 | Gene names | EIF3S2, TRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 2 (eIF-3 beta) (eIF3 p36) (eIF3i) (TGF-beta receptor-interacting protein 1) (TRIP-1). | |||||
|
IF32_MOUSE
|
||||||
| NC score | 0.010656 (rank : 171) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZD9 | Gene names | Eif3s2, Trip1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 2 (eIF-3 beta) (eIF3 p36) (eIF3i) (TGF-beta receptor-interacting protein 1) (TRIP-1). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.010538 (rank : 172) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
CBL_HUMAN
|
||||||
| NC score | 0.009947 (rank : 173) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P22681 | Gene names | CBL, CBL2, RNF55 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene) (RING finger protein 55). | |||||
|
SP5_HUMAN
|
||||||
| NC score | 0.009372 (rank : 174) | θ value | 0.163984 (rank : 35) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
CAR10_MOUSE
|
||||||
| NC score | 0.009268 (rank : 175) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P58660 | Gene names | Card10, Bimp1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 10 (Bcl10-interacting MAGUK protein 1) (Bimp1). | |||||
|
FLNC_HUMAN
|
||||||
| NC score | 0.009085 (rank : 176) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14315, O95303, Q07985, Q9NS12, Q9NYE5, Q9UMR8, Q9Y503 | Gene names | FLNC, ABPL, FLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-C (Gamma-filamin) (Filamin-2) (Protein FLNc) (Actin-binding- like protein) (ABP-L) (ABP-280-like protein). | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | 0.008287 (rank : 177) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
SC6A5_MOUSE
|
||||||
| NC score | 0.008089 (rank : 178) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q761V0, Q8CFM5, Q91ZQ2 | Gene names | Slc6a5, Glyt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.007165 (rank : 179) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
CAN9_HUMAN
|
||||||
| NC score | 0.006306 (rank : 180) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14815, Q9NS74 | Gene names | CAPN9, NCL4 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-9 (EC 3.4.22.-) (Digestive tract-specific calpain) (nCL-4) (CG36 protein). | |||||
|
ULK1_MOUSE
|
||||||
| NC score | 0.004089 (rank : 181) | θ value | 0.365318 (rank : 42) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
MK15_HUMAN
|
||||||
| NC score | 0.003749 (rank : 182) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TD08, Q2TCF9, Q8N362 | Gene names | MAPK15, ERK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 8). | |||||
|
PAK4_MOUSE
|
||||||
| NC score | 0.003727 (rank : 183) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1098 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BTW9, Q6ZPX0, Q80Z97, Q9CS71 | Gene names | Pak4, Kiaa1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||
|
CELR3_MOUSE
|
||||||
| NC score | 0.002736 (rank : 184) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
PAK7_MOUSE
|
||||||
| NC score | 0.001264 (rank : 185) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C015, Q3TQJ7, Q6RWS7 | Gene names | Pak7, Pak5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PAK 7 (EC 2.7.11.1) (p21-activated kinase 7) (PAK-7) (p21-activated kinase 5). | |||||
|
MK04_MOUSE
|
||||||
| NC score | 0.001085 (rank : 186) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6P5G0 | Gene names | Mapk4, Erk4, Prkm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 4 (EC 2.7.11.24) (Extracellular signal-regulated kinase 4) (ERK-4). | |||||
|
ZFP13_MOUSE
|
||||||
| NC score | -0.001630 (rank : 187) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10754, Q80WS0, Q8CDD0 | Gene names | Zfp13, Krox-8, Zfp-13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 13 (Zfp-13) (Krox-8 protein). | |||||