Please be patient as the page loads
|
B4GT3_HUMAN
|
||||||
| SwissProt Accessions | O60512, O60910, Q9BPZ4, Q9H8T2 | Gene names | B4GALT3 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
B4GT3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O60512, O60910, Q9BPZ4, Q9H8T2 | Gene names | B4GALT3 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998612 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91YY2, Q9QY13 | Gene names | B4galt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT1_HUMAN
|
||||||
| θ value | 4.20666e-90 (rank : 3) | NC score | 0.965199 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT1_MOUSE
|
||||||
| θ value | 5.14231e-88 (rank : 4) | NC score | 0.976128 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15535 | Gene names | B4galt1, Ggtb, Ggtb2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT4_MOUSE
|
||||||
| θ value | 3.81361e-83 (rank : 5) | NC score | 0.981515 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JJ04, Q8BR54, Q9QY12 | Gene names | B4galt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT4_HUMAN
|
||||||
| θ value | 3.22841e-82 (rank : 6) | NC score | 0.980658 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60513, Q68D68, Q9BSW3, Q9C078 | Gene names | B4GALT4 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT2_HUMAN
|
||||||
| θ value | 7.95186e-81 (rank : 7) | NC score | 0.974501 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60909, O60511, Q5T4X5, Q5T4Y5, Q9BUP6, Q9NSY7 | Gene names | B4GALT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT2_MOUSE
|
||||||
| θ value | 8.79181e-80 (rank : 8) | NC score | 0.975512 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z2Y2, Q3TMP2 | Gene names | B4galt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT5_HUMAN
|
||||||
| θ value | 1.77973e-64 (rank : 9) | NC score | 0.954198 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43286, Q2M394, Q9UJQ8 | Gene names | B4GALT5 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT6_HUMAN
|
||||||
| θ value | 2.56992e-63 (rank : 10) | NC score | 0.952626 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UBX8, O60514 | Gene names | B4GALT6 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT6_MOUSE
|
||||||
| θ value | 2.17557e-62 (rank : 11) | NC score | 0.951562 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WVK5 | Gene names | B4galt6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT5_MOUSE
|
||||||
| θ value | 4.24587e-58 (rank : 12) | NC score | 0.952400 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JMK0 | Gene names | B4galt5, Bgt-5 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT7_MOUSE
|
||||||
| θ value | 6.61148e-27 (rank : 13) | NC score | 0.882051 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R087, Q7TNU3 | Gene names | B4galt7, Xgalt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
B4GT7_HUMAN
|
||||||
| θ value | 1.12775e-26 (rank : 14) | NC score | 0.875093 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UBV7, Q9UHN2 | Gene names | B4GALT7, XGALT1 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
GALT3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.039847 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14435, Q7Z476 | Gene names | GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 3) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 3) (Polypeptide GalNAc transferase 3) (GalNAc-T3) (pp-GaNTase 3). | |||||
|
EMID1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.024054 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91VF5 | Gene names | Emid1, Emu1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EMI domain-containing protein 1 precursor (Protein Emu1) (Emilin and multimerin domain-containing protein 1). | |||||
|
GALT6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.038951 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NCL4, Q8IYH4, Q9H6G2, Q9UIV5 | Gene names | GALNT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
GALT6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.042522 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C7U7, Q3TWF0, Q8CED2, Q9QZ16 | Gene names | Galnt6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
COCA1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.017208 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60847, P70322 | Gene names | Col12a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
GALT3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.035608 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70419, Q80V55 | Gene names | Galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 3) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 3) (Polypeptide GalNAc transferase 3) (GalNAc-T3) (pp-GaNTase 3). | |||||
|
COEA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.019908 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q05707, O00260, O00261, O00262, Q05708, Q5XJ18, Q96C67 | Gene names | COL14A1, UND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor (Undulin). | |||||
|
ADA12_MOUSE
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.009774 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
COEA1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.018567 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80X19, Q8C6X3, Q9WV05 | Gene names | Col14a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor. | |||||
|
COIA1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.020594 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
ADA12_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.007120 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43184, O60470 | Gene names | ADAM12, MLTN | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
BAT3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.028288 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
CHSS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.117933 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86X52, Q6UX38, Q7LFU5, Q9Y2J5 | Gene names | CHSY1, CHSY, CSS1, KIAA0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase 1) (Chondroitin sulfate synthase 1) (N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.111615 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
COHA1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.020657 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
CHSS3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.091715 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
COIA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.020292 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
CGAT2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.052857 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C1F4, Q6PAP6, Q8R5A2, Q9D2M1 | Gene names | Galnact2, Chgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CO3A1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.015123 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
COBA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.018362 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P13942, Q07751, Q13271, Q13272, Q13273, Q99866, Q9UIP9 | Gene names | COL11A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
COBA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.018124 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
COHA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.018489 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UMD9, Q02802, Q99018, Q9NQK9, Q9UC14 | Gene names | COL17A1, BP180, BPAG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.005077 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.011937 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PTC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.015821 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35595, Q546T0 | Gene names | Ptch2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 2 (PTC2). | |||||
|
WNT16_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.005305 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYS1 | Gene names | Wnt16 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Wnt-16 precursor. | |||||
|
CO1A1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.015109 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
GA2L1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.009282 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99501, Q53EN7, Q92640, Q9BUY9 | Gene names | GAS2L1, GAR22 | |||
|
Domain Architecture |

|
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1) (GAS2-related protein on chromosome 22) (GAR22 protein). | |||||
|
GALT7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.017567 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80VA0, Q8BY62, Q8BZ70, Q8BZW0, Q91VW6, Q99MD7 | Gene names | Galnt7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylgalactosaminyltransferase 7 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 7) (UDP-GalNAc:polypeptide N- acetylgalactosaminyltransferase 7) (Polypeptide GalNAc transferase 7) (GalNAc-T7) (pp-GaNTase 7). | |||||
|
PTC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.014545 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6C5, O95341, O95856, Q5QP87, Q6UX14 | Gene names | PTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 2 (PTC2). | |||||
|
B4GN4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.064716 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
BAT3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.020668 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
CO2A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.016430 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P02458, Q12985, Q14009, Q14044, Q14046, Q14056, Q14058, Q16672, Q6LBY1, Q6LBY2, Q6LBY3, Q99227, Q9UE38, Q9UE39, Q9UE40, Q9UE41, Q9UE42, Q9UE43 | Gene names | COL2A1 | |||
|
Domain Architecture |
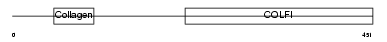
|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
CO2A1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.015550 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P28481 | Gene names | Col2a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
EMID1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.018983 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96A84, Q6ICG1, Q86SS7 | Gene names | EMID1, EMU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EMI domain-containing protein 1 precursor (Protein Emu1) (Emilin and multimerin domain-containing protein 1). | |||||
|
GLT11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.016588 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NCW6, Q6PCD1, Q9H6C2, Q9H6Z5, Q9UDR8 | Gene names | GALNT11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 11 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 11) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 11) (Polypeptide GalNAc transferase 11) (GalNAc-T11) (pp-GaNTase 11). | |||||
|
B4GN4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.053495 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
CGAT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.051643 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BJQ9, Q3UZX6, Q8BWV9, Q8BZU7, Q8C195, Q8R0M6 | Gene names | Galnact1, Chgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
B4GT3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O60512, O60910, Q9BPZ4, Q9H8T2 | Gene names | B4GALT3 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT3_MOUSE
|
||||||
| NC score | 0.998612 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91YY2, Q9QY13 | Gene names | B4galt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT4_MOUSE
|
||||||
| NC score | 0.981515 (rank : 3) | θ value | 3.81361e-83 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JJ04, Q8BR54, Q9QY12 | Gene names | B4galt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT4_HUMAN
|
||||||
| NC score | 0.980658 (rank : 4) | θ value | 3.22841e-82 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60513, Q68D68, Q9BSW3, Q9C078 | Gene names | B4GALT4 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT1_MOUSE
|
||||||
| NC score | 0.976128 (rank : 5) | θ value | 5.14231e-88 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15535 | Gene names | B4galt1, Ggtb, Ggtb2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT2_MOUSE
|
||||||
| NC score | 0.975512 (rank : 6) | θ value | 8.79181e-80 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z2Y2, Q3TMP2 | Gene names | B4galt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT2_HUMAN
|
||||||
| NC score | 0.974501 (rank : 7) | θ value | 7.95186e-81 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60909, O60511, Q5T4X5, Q5T4Y5, Q9BUP6, Q9NSY7 | Gene names | B4GALT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT1_HUMAN
|
||||||
| NC score | 0.965199 (rank : 8) | θ value | 4.20666e-90 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT5_HUMAN
|
||||||
| NC score | 0.954198 (rank : 9) | θ value | 1.77973e-64 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43286, Q2M394, Q9UJQ8 | Gene names | B4GALT5 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT6_HUMAN
|
||||||
| NC score | 0.952626 (rank : 10) | θ value | 2.56992e-63 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UBX8, O60514 | Gene names | B4GALT6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT5_MOUSE
|
||||||
| NC score | 0.952400 (rank : 11) | θ value | 4.24587e-58 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JMK0 | Gene names | B4galt5, Bgt-5 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT6_MOUSE
|
||||||
| NC score | 0.951562 (rank : 12) | θ value | 2.17557e-62 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WVK5 | Gene names | B4galt6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT7_MOUSE
|
||||||
| NC score | 0.882051 (rank : 13) | θ value | 6.61148e-27 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R087, Q7TNU3 | Gene names | B4galt7, Xgalt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
B4GT7_HUMAN
|
||||||
| NC score | 0.875093 (rank : 14) | θ value | 1.12775e-26 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UBV7, Q9UHN2 | Gene names | B4GALT7, XGALT1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 7 (EC 2.4.1.-) (Beta-1,4-GalTase 7) (Beta4Gal-T7) (b4Gal-T7) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 7) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 7) [Includes: Xylosylprotein 4-beta- galactosyltransferase (EC 2.4.1.133) (UDP-galactose:beta-xylose beta- 1,4-galactosyltransferase) (Xylosylprotein beta-1,4- galactosyltransferase) (XGPT) (Proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I) (XGalT-1)]. | |||||
|
CHSS1_HUMAN
|
||||||
| NC score | 0.117933 (rank : 15) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86X52, Q6UX38, Q7LFU5, Q9Y2J5 | Gene names | CHSY1, CHSY, CSS1, KIAA0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase 1) (Chondroitin sulfate synthase 1) (N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS1_MOUSE
|
||||||
| NC score | 0.111615 (rank : 16) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS3_HUMAN
|
||||||
| NC score | 0.091715 (rank : 17) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
B4GN4_HUMAN
|
||||||
| NC score | 0.064716 (rank : 18) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GN4_MOUSE
|
||||||
| NC score | 0.053495 (rank : 19) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
CGAT2_MOUSE
|
||||||
| NC score | 0.052857 (rank : 20) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C1F4, Q6PAP6, Q8R5A2, Q9D2M1 | Gene names | Galnact2, Chgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CGAT1_MOUSE
|
||||||
| NC score | 0.051643 (rank : 21) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BJQ9, Q3UZX6, Q8BWV9, Q8BZU7, Q8C195, Q8R0M6 | Gene names | Galnact1, Chgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
GALT6_MOUSE
|
||||||
| NC score | 0.042522 (rank : 22) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C7U7, Q3TWF0, Q8CED2, Q9QZ16 | Gene names | Galnt6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
GALT3_HUMAN
|
||||||
| NC score | 0.039847 (rank : 23) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14435, Q7Z476 | Gene names | GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 3) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 3) (Polypeptide GalNAc transferase 3) (GalNAc-T3) (pp-GaNTase 3). | |||||
|
GALT6_HUMAN
|
||||||
| NC score | 0.038951 (rank : 24) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NCL4, Q8IYH4, Q9H6G2, Q9UIV5 | Gene names | GALNT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
GALT3_MOUSE
|
||||||
| NC score | 0.035608 (rank : 25) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70419, Q80V55 | Gene names | Galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 3) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 3) (Polypeptide GalNAc transferase 3) (GalNAc-T3) (pp-GaNTase 3). | |||||
|
BAT3_HUMAN
|
||||||
| NC score | 0.028288 (rank : 26) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
EMID1_MOUSE
|
||||||
| NC score | 0.024054 (rank : 27) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91VF5 | Gene names | Emid1, Emu1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EMI domain-containing protein 1 precursor (Protein Emu1) (Emilin and multimerin domain-containing protein 1). | |||||
|
BAT3_MOUSE
|
||||||
| NC score | 0.020668 (rank : 28) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
COHA1_MOUSE
|
||||||
| NC score | 0.020657 (rank : 29) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
COIA1_MOUSE
|
||||||
| NC score | 0.020594 (rank : 30) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
COIA1_HUMAN
|
||||||
| NC score | 0.020292 (rank : 31) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
COEA1_HUMAN
|
||||||
| NC score | 0.019908 (rank : 32) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q05707, O00260, O00261, O00262, Q05708, Q5XJ18, Q96C67 | Gene names | COL14A1, UND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor (Undulin). | |||||
|
EMID1_HUMAN
|
||||||
| NC score | 0.018983 (rank : 33) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96A84, Q6ICG1, Q86SS7 | Gene names | EMID1, EMU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EMI domain-containing protein 1 precursor (Protein Emu1) (Emilin and multimerin domain-containing protein 1). | |||||
|
COEA1_MOUSE
|
||||||
| NC score | 0.018567 (rank : 34) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80X19, Q8C6X3, Q9WV05 | Gene names | Col14a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor. | |||||
|
COHA1_HUMAN
|
||||||
| NC score | 0.018489 (rank : 35) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UMD9, Q02802, Q99018, Q9NQK9, Q9UC14 | Gene names | COL17A1, BP180, BPAG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
COBA2_HUMAN
|
||||||
| NC score | 0.018362 (rank : 36) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P13942, Q07751, Q13271, Q13272, Q13273, Q99866, Q9UIP9 | Gene names | COL11A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
COBA2_MOUSE
|
||||||
| NC score | 0.018124 (rank : 37) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
GALT7_MOUSE
|
||||||
| NC score | 0.017567 (rank : 38) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80VA0, Q8BY62, Q8BZ70, Q8BZW0, Q91VW6, Q99MD7 | Gene names | Galnt7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylgalactosaminyltransferase 7 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 7) (UDP-GalNAc:polypeptide N- acetylgalactosaminyltransferase 7) (Polypeptide GalNAc transferase 7) (GalNAc-T7) (pp-GaNTase 7). | |||||
|
COCA1_MOUSE
|
||||||
| NC score | 0.017208 (rank : 39) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60847, P70322 | Gene names | Col12a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
GLT11_HUMAN
|
||||||
| NC score | 0.016588 (rank : 40) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NCW6, Q6PCD1, Q9H6C2, Q9H6Z5, Q9UDR8 | Gene names | GALNT11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 11 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 11) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 11) (Polypeptide GalNAc transferase 11) (GalNAc-T11) (pp-GaNTase 11). | |||||
|
CO2A1_HUMAN
|
||||||
| NC score | 0.016430 (rank : 41) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P02458, Q12985, Q14009, Q14044, Q14046, Q14056, Q14058, Q16672, Q6LBY1, Q6LBY2, Q6LBY3, Q99227, Q9UE38, Q9UE39, Q9UE40, Q9UE41, Q9UE42, Q9UE43 | Gene names | COL2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
PTC2_MOUSE
|
||||||
| NC score | 0.015821 (rank : 42) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35595, Q546T0 | Gene names | Ptch2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 2 (PTC2). | |||||
|
CO2A1_MOUSE
|
||||||
| NC score | 0.015550 (rank : 43) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P28481 | Gene names | Col2a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
CO3A1_MOUSE
|
||||||
| NC score | 0.015123 (rank : 44) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
CO1A1_HUMAN
|
||||||
| NC score | 0.015109 (rank : 45) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
PTC2_HUMAN
|
||||||
| NC score | 0.014545 (rank : 46) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6C5, O95341, O95856, Q5QP87, Q6UX14 | Gene names | PTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 2 (PTC2). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.011937 (rank : 47) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
ADA12_MOUSE
|
||||||
| NC score | 0.009774 (rank : 48) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
GA2L1_HUMAN
|
||||||
| NC score | 0.009282 (rank : 49) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99501, Q53EN7, Q92640, Q9BUY9 | Gene names | GAS2L1, GAR22 | |||
|
Domain Architecture |

|
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1) (GAS2-related protein on chromosome 22) (GAR22 protein). | |||||
|
ADA12_HUMAN
|
||||||
| NC score | 0.007120 (rank : 50) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43184, O60470 | Gene names | ADAM12, MLTN | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
WNT16_MOUSE
|
||||||
| NC score | 0.005305 (rank : 51) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYS1 | Gene names | Wnt16 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Wnt-16 precursor. | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.005077 (rank : 52) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||