Please be patient as the page loads
|
B4GN4_MOUSE
|
||||||
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
B4GN3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.931433 (rank : 3) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6L9W6, Q6ZNC1, Q8N7T6 | Gene names | B4GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
B4GN4_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.962859 (rank : 2) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GN4_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 129 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GN3_MOUSE
|
||||||
| θ value | 1.99884e-124 (rank : 4) | NC score | 0.925262 (rank : 4) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6L8S8 | Gene names | B4galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
CGAT1_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 5) | NC score | 0.578573 (rank : 5) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TDX6, Q6P9G6, Q8IUF9, Q9NSQ7, Q9NUM9 | Gene names | GALNACT1, CHGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT1_MOUSE
|
||||||
| θ value | 8.95645e-16 (rank : 6) | NC score | 0.575668 (rank : 6) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BJQ9, Q3UZX6, Q8BWV9, Q8BZU7, Q8C195, Q8R0M6 | Gene names | Galnact1, Chgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT2_HUMAN
|
||||||
| θ value | 1.16975e-15 (rank : 7) | NC score | 0.562996 (rank : 7) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8N6G5, Q6MZJ5, Q6MZP6, Q8TCH4, Q9P1I6 | Gene names | GALNACT2, CHGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CGAT2_MOUSE
|
||||||
| θ value | 3.76295e-14 (rank : 8) | NC score | 0.555996 (rank : 8) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C1F4, Q6PAP6, Q8R5A2, Q9D2M1 | Gene names | Galnact2, Chgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CHSS1_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 9) | NC score | 0.491343 (rank : 9) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86X52, Q6UX38, Q7LFU5, Q9Y2J5 | Gene names | CHSY1, CHSY, CSS1, KIAA0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase 1) (Chondroitin sulfate synthase 1) (N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS1_MOUSE
|
||||||
| θ value | 6.00763e-12 (rank : 10) | NC score | 0.488152 (rank : 10) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS3_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 11) | NC score | 0.470132 (rank : 11) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
CHSS2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 12) | NC score | 0.229030 (rank : 12) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZ52, Q6UXD6, Q7L4G1, Q9H0F8, Q9H618 | Gene names | CSS2, CHPF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase) (Chondroitin-polymerizing factor). | |||||
|
CHSS2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.228838 (rank : 13) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6IQX7 | Gene names | Css2, D1Bwg1363e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 14) | NC score | 0.046006 (rank : 71) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
B4GT6_HUMAN
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.092463 (rank : 18) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UBX8, O60514 | Gene names | B4GALT6 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT6_MOUSE
|
||||||
| θ value | 0.125558 (rank : 16) | NC score | 0.092022 (rank : 19) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WVK5 | Gene names | B4galt6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
GALT3_MOUSE
|
||||||
| θ value | 0.125558 (rank : 17) | NC score | 0.092676 (rank : 17) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70419, Q80V55 | Gene names | Galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 3) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 3) (Polypeptide GalNAc transferase 3) (GalNAc-T3) (pp-GaNTase 3). | |||||
|
LTBP3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.012799 (rank : 126) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NS15, O15107, Q96HB9, Q9H7K2, Q9UFN4 | Gene names | LTBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
WASP_MOUSE
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.069442 (rank : 41) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
B4GT5_HUMAN
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.086222 (rank : 23) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43286, Q2M394, Q9UJQ8 | Gene names | B4GALT5 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT5_MOUSE
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.086699 (rank : 22) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JMK0 | Gene names | B4galt5, Bgt-5 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
GALT3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.090650 (rank : 20) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14435, Q7Z476 | Gene names | GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 3) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 3) (Polypeptide GalNAc transferase 3) (GalNAc-T3) (pp-GaNTase 3). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.040600 (rank : 75) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.036196 (rank : 79) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
WASP_HUMAN
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.067740 (rank : 42) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
B4GT2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.110995 (rank : 15) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60909, O60511, Q5T4X5, Q5T4Y5, Q9BUP6, Q9NSY7 | Gene names | B4GALT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
EFS_HUMAN
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.036037 (rank : 81) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
GALT6_HUMAN
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.089674 (rank : 21) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NCL4, Q8IYH4, Q9H6G2, Q9UIV5 | Gene names | GALNT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.038872 (rank : 76) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.033665 (rank : 83) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.040725 (rank : 74) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
WASL_MOUSE
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.063915 (rank : 47) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
B4GT2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.109475 (rank : 16) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z2Y2, Q3TMP2 | Gene names | B4galt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
CHGUT_HUMAN
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.196042 (rank : 14) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2E5, Q6P2I4, Q6UXD2 | Gene names | CSGLCAT, KIAA1402 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate glucuronyltransferase (EC 2.4.1.226) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (Chondroitin glucuronyltransferase II) (CSGlcA-T). | |||||
|
LR37B_HUMAN
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.031720 (rank : 85) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
SSH3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.019490 (rank : 106) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TE77, Q6PK42, Q76I75, Q8N9L8, Q8WYL0, Q9NV45, Q9NWZ7 | Gene names | SSH3, SSH3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (hSSH-3L). | |||||
|
PDE1C_HUMAN
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.014098 (rank : 121) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
CSPG5_MOUSE
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.034758 (rank : 82) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q71M36, Q71M37, Q7TNT8, Q8BPJ5, Q9QY32 | Gene names | Cspg5, Caleb, Ngc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
GALT6_MOUSE
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.084218 (rank : 24) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8C7U7, Q3TWF0, Q8CED2, Q9QZ16 | Gene names | Galnt6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.018894 (rank : 107) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
RHG07_MOUSE
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.015586 (rank : 117) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.044531 (rank : 72) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
GLT13_HUMAN
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.075481 (rank : 33) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IUC8, Q6ZWG1, Q96PX0, Q9UIE5 | Gene names | GALNT13, KIAA1918 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 13 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 13) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 13) (Polypeptide GalNAc transferase 13) (GalNAc-T13) (pp-GaNTase 13). | |||||
|
GLT13_MOUSE
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.075492 (rank : 32) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CF93, Q8BLE4, Q8BYT3 | Gene names | Galnt13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 13 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 13) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 13) (Polypeptide GalNAc transferase 13) (GalNAc-T13) (pp-GaNTase 13). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.004695 (rank : 142) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.021951 (rank : 98) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
GLT12_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.077712 (rank : 25) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BGT9 | Gene names | Galnt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 12 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 12) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 12) (Polypeptide GalNAc transferase 12) (GalNAc-T12) (pp-GaNTase 12). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.028383 (rank : 89) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.050830 (rank : 70) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.055822 (rank : 64) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
FGD1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.016015 (rank : 113) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
GALT4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.076817 (rank : 26) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N4A0, O00208 | Gene names | GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 4) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 4) (Polypeptide GalNAc transferase 4) (GalNAc-T4) (pp-GaNTase 4). | |||||
|
GALT4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.076806 (rank : 27) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O08832, Q3U681 | Gene names | Galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 4) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 4) (Polypeptide GalNAc transferase 4) (GalNAc-T4) (pp-GaNTase 4). | |||||
|
GLTL1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.075825 (rank : 31) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N428, Q9ULT9 | Gene names | GALNTL1, KIAA1130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 1 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 1) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 1) (Polypeptide GalNAc transferase-like protein 1) (GalNAc-T-like protein 1) (pp-GaNTase-like protein 1). | |||||
|
GLTL1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.075844 (rank : 30) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JJ61 | Gene names | Galntl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 1 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 1) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 1) (Polypeptide GalNAc transferase-like protein 1) (GalNAc-T-like protein 1) (pp-GaNTase-like protein 1). | |||||
|
SON_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.042300 (rank : 73) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
T240L_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.029540 (rank : 87) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q71F56, Q68DN4, Q9H8C0, Q9NSY9, Q9UFD8, Q9UPX5 | Gene names | THRAP2, KIAA1025, PROSIT240, TRAP240L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
COIA1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.013062 (rank : 124) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
GALT5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.075024 (rank : 34) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8C102 | Gene names | Galnt5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
GLT14_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.075908 (rank : 28) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96FL9, Q8IVI4, Q9BRH1, Q9H827, Q9H9J8 | Gene names | GALNT14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 14 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 14) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 14) (Polypeptide GalNAc transferase 14) (GalNAc-T14) (pp-GaNTase 14). | |||||
|
GLT14_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.075869 (rank : 29) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BVG5, Q60GT2, Q8BTI6, Q9DCJ2 | Gene names | Galnt14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 14 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 14) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 14) (Polypeptide GalNAc transferase 14) (GalNAc-T14) (pp-GaNTase 14). | |||||
|
K1949_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.026842 (rank : 90) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6NYC8, Q68CK8, Q6ZUJ6, Q8NDQ4, Q8TF52, Q9BRL9 | Gene names | KIAA1949 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949. | |||||
|
LEUK_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.033344 (rank : 84) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15702 | Gene names | Spn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Ly- 48) (B cell differentiation antigen LP-3) (CD43 antigen). | |||||
|
POGZ_MOUSE
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.003948 (rank : 143) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BZH4, Q4VA94, Q80TZ8, Q8C0K1, Q8K294 | Gene names | Pogz, Kiaa0461 | |||
|
Domain Architecture |

|
|||||
| Description | Pogo transposable element with ZNF domain. | |||||
|
DYN3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.012873 (rank : 125) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
GA2L2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.038373 (rank : 77) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
GALT1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.071039 (rank : 37) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q10472, Q86TJ7, Q9UM86 | Gene names | GALNT1 | |||
|
Domain Architecture |

|
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 1 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 1) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 1) (Polypeptide GalNAc transferase 1) (GalNAc-T1) (pp-GaNTase 1) [Contains: Polypeptide N-acetylgalactosaminyltransferase 1 soluble form]. | |||||
|
GALT1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.071009 (rank : 38) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08912, Q5BKS3, Q7TND1 | Gene names | Galnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 1 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 1) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 1) (Polypeptide GalNAc transferase 1) (GalNAc-T1) (pp-GaNTase 1) [Contains: Polypeptide N-acetylgalactosaminyltransferase 1 soluble form]. | |||||
|
GALT5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.073879 (rank : 36) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z7M9, Q9UGK7, Q9UHL6 | Gene names | GALNT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
LEG3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.015946 (rank : 114) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P16110 | Gene names | Lgals3 | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-34 galactoside-binding lectin). | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.010047 (rank : 131) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
RTN4R_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.006141 (rank : 140) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BZR6 | Gene names | RTN4R, NOGOR | |||
|
Domain Architecture |
|
|||||
| Description | Reticulon-4 receptor precursor (Nogo receptor) (NgR) (Nogo-66 receptor). | |||||
|
SC6A5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.008699 (rank : 135) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q761V0, Q8CFM5, Q91ZQ2 | Gene names | Slc6a5, Glyt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
WASL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.054776 (rank : 66) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
BSCL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.020865 (rank : 102) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
DIAP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.020113 (rank : 105) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
FGD1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.014877 (rank : 119) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
FMN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.023113 (rank : 94) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
GLT12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.074664 (rank : 35) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IXK2, Q5TCF7, Q8NG54, Q96CT9, Q9H771 | Gene names | GALNT12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 12 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 12) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 12) (Polypeptide GalNAc transferase 12) (GalNAc-T12) (pp-GaNTase 12). | |||||
|
LATS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.002960 (rank : 145) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
MICA3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.008480 (rank : 136) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7RTP6, Q6ICK4, Q9P2I3 | Gene names | MICAL3, KIAA1364 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-3. | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.002538 (rank : 149) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
RUSC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.037270 (rank : 78) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BG26, Q6PHT9 | Gene names | Rusc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1. | |||||
|
3BP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.036046 (rank : 80) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
APC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.015723 (rank : 116) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
FMNL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.024922 (rank : 93) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
GALT2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.070121 (rank : 39) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q10471, Q9NPY4 | Gene names | GALNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 2 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 2) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 2) (Polypeptide GalNAc transferase 2) (GalNAc-T2) (pp-GaNTase 2) [Contains: Polypeptide N-acetylgalactosaminyltransferase 2 soluble form]. | |||||
|
GALT2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.069917 (rank : 40) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PB93, Q7TSI5, Q8BL27, Q922K5, Q99ME1 | Gene names | Galnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 2 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 2) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 2) (Polypeptide GalNAc transferase 2) (GalNAc-T2) (pp-GaNTase 2) [Contains: Polypeptide N-acetylgalactosaminyltransferase 2 soluble form]. | |||||
|
LATS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.002691 (rank : 148) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95835, Q6PKD0 | Gene names | LATS1, WARTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase) (h-warts). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.030211 (rank : 86) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.003672 (rank : 144) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TPP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.017998 (rank : 110) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14773, Q53HT1, Q5JAK6, Q6UX56, Q96C37 | Gene names | TPP1, CLN2, GIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Tripeptidyl-peptidase 1 precursor (EC 3.4.14.9) (Tripeptidyl-peptidase I) (TPP-I) (Tripeptidyl aminopeptidase) (Lysosomal pepstatin insensitive protease) (LPIC) (Growth-inhibiting gene 1 protein). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.026336 (rank : 92) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
CPSF6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.021117 (rank : 101) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CPSF6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.021218 (rank : 100) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
FOG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.002716 (rank : 147) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
S2A4R_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.022261 (rank : 97) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NR83, Q6F6I6, Q6F6I7, Q6GTK5, Q8TAH5, Q8WVW7, Q96QD3, Q9BV85 | Gene names | SLC2A4RG, HDBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SLC2A4 regulator (GLUT4 enhancer factor) (GEF) (Huntington disease gene regulatory region-binding protein 1) (HDBP-1). | |||||
|
TMUB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.015120 (rank : 118) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3V209, Q8K0X0, Q91YQ0, Q9D8C5 | Gene names | Tmub2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 2. | |||||
|
WASF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.015756 (rank : 115) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.017993 (rank : 111) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.009430 (rank : 133) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.018052 (rank : 109) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.009827 (rank : 132) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
ESR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.002810 (rank : 146) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P03372, Q13511, Q14276, Q9NU51, Q9UDZ7, Q9UIS7 | Gene names | ESR1, ESR, NR3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen receptor (ER) (Estradiol receptor) (ER-alpha). | |||||
|
ETV3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.006730 (rank : 138) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.014283 (rank : 120) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
IF32_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.011773 (rank : 129) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13347 | Gene names | EIF3S2, TRIP1 | |||
|
Domain Architecture |
|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 2 (eIF-3 beta) (eIF3 p36) (eIF3i) (TGF-beta receptor-interacting protein 1) (TRIP-1). | |||||
|
IF32_MOUSE
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.011771 (rank : 130) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZD9 | Gene names | Eif3s2, Trip1 | |||
|
Domain Architecture |
|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 2 (eIF-3 beta) (eIF3 p36) (eIF3i) (TGF-beta receptor-interacting protein 1) (TRIP-1). | |||||
|
KIF3C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.002411 (rank : 150) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14782, O43544 | Gene names | KIF3C | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
LIPB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.006322 (rank : 139) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35711, Q7TMG4, Q8CBS6, Q99KX6 | Gene names | Ppfibp2, Cclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2) (Coiled-coil-like protein 1). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.026378 (rank : 91) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
PRP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.018169 (rank : 108) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.016282 (rank : 112) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
TM108_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.021823 (rank : 99) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
WASIP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.022761 (rank : 95) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
CO039_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.028393 (rank : 88) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
FMNL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.020135 (rank : 104) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JL26, Q6KAN4, Q9Z2V7 | Gene names | Fmnl1, Frl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-like protein 1 (Formin-related protein). | |||||
|
GLT10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.066929 (rank : 44) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86SR1, Q6IN56, Q86VP8, Q8IXJ2, Q8TEJ2, Q96IV2, Q9H8E1, Q9Y4M4 | Gene names | GALNT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 10 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 10) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 10) (Polypeptide GalNAc transferase 10) (GalNAc-T10) (pp-GaNTase 10). | |||||
|
GLT10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.066885 (rank : 45) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6P9S7, Q6KAQ2, Q8BZU8, Q91YJ6 | Gene names | Galnt10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 10 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 10) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 10) (Polypeptide GalNAc transferase 10) (GalNAc-T10) (pp-GaNTase 10). | |||||
|
IASPP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.009233 (rank : 134) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
JHD3B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.005017 (rank : 141) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
NEBL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.007996 (rank : 137) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O76041, Q9UIC4 | Gene names | NEBL | |||
|
Domain Architecture |

|
|||||
| Description | Nebulette (Actin-binding Z-disk protein). | |||||
|
PAK7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.000238 (rank : 151) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C015, Q3TQJ7, Q6RWS7 | Gene names | Pak7, Pak5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PAK 7 (EC 2.7.11.1) (p21-activated kinase 7) (PAK-7) (p21-activated kinase 5). | |||||
|
PCX1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.012177 (rank : 128) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96RV3, O94897, Q96AI7, Q9Y2J9 | Gene names | PCNX, KIAA0805, KIAA0995, PCNXL1 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 1 (Pecanex homolog). | |||||
|
SFR15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.020667 (rank : 103) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.013641 (rank : 123) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.013831 (rank : 122) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
TNFL6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.012286 (rank : 127) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48023, Q9BZP9 | Gene names | FASLG, APT1LG1, FASL, TNFSF6 | |||
|
Domain Architecture |
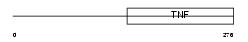
|
|||||
| Description | Tumor necrosis factor ligand superfamily member 6 (Fas antigen ligand) (Fas ligand) (CD178 antigen) (CD95L protein) (Apoptosis antigen ligand) (APTL) [Contains: Tumor necrosis factor ligand superfamily member 6, membrane form; Tumor necrosis factor ligand superfamily member 6, soluble form]. | |||||
|
WASIP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.022374 (rank : 96) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
B3GLT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.051502 (rank : 69) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6Y288, Q5W0H2, Q6NUI3 | Gene names | B3GALTL, B3GTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
B3GLT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.055562 (rank : 65) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BHT6, Q0PCR7, Q149S5 | Gene names | B3galtl, Gm1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
B4GT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.062922 (rank : 52) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.067415 (rank : 43) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P15535 | Gene names | B4galt1, Ggtb, Ggtb2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.053495 (rank : 68) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60512, O60910, Q9BPZ4, Q9H8T2 | Gene names | B4GALT3 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.053814 (rank : 67) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91YY2, Q9QY13 | Gene names | B4galt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.060232 (rank : 59) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60513, Q68D68, Q9BSW3, Q9C078 | Gene names | B4GALT4 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.061981 (rank : 56) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JJ04, Q8BR54, Q9QY12 | Gene names | B4galt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
GALT7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.062788 (rank : 54) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86SF2, Q7Z5W7, Q9UJ28 | Gene names | GALNT7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylgalactosaminyltransferase 7 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 7) (UDP-GalNAc:polypeptide N- acetylgalactosaminyltransferase 7) (Polypeptide GalNAc transferase 7) (GalNAc-T7) (pp-GaNTase 7). | |||||
|
GALT7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.062962 (rank : 51) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q80VA0, Q8BY62, Q8BZ70, Q8BZW0, Q91VW6, Q99MD7 | Gene names | Galnt7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylgalactosaminyltransferase 7 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 7) (UDP-GalNAc:polypeptide N- acetylgalactosaminyltransferase 7) (Polypeptide GalNAc transferase 7) (GalNAc-T7) (pp-GaNTase 7). | |||||
|
GALT8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.060299 (rank : 58) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NY28 | Gene names | GALNT8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable polypeptide N-acetylgalactosaminyltransferase 8 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 8) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 8) (Polypeptide GalNAc transferase 8) (GalNAc-T8) (pp-GaNTase 8). | |||||
|
GALT9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.060501 (rank : 57) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HCQ5, Q6NT54, Q8NFR1 | Gene names | GALNT9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 9 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 9) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 9) (Polypeptide GalNAc transferase 9) (GalNAc-T9) (pp-GaNTase 9). | |||||
|
GLT11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.063271 (rank : 49) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NCW6, Q6PCD1, Q9H6C2, Q9H6Z5, Q9UDR8 | Gene names | GALNT11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 11 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 11) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 11) (Polypeptide GalNAc transferase 11) (GalNAc-T11) (pp-GaNTase 11). | |||||
|
GLT11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.063224 (rank : 50) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q921L8, Q3TT83, Q8BU26, Q8K032 | Gene names | Galnt11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 11 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 11) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 11) (Polypeptide GalNAc transferase 11) (GalNAc-T11) (pp-GaNTase 11). | |||||
|
GLTL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.063976 (rank : 46) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N3T1, Q86T60, Q96C46, Q96DJ5 | Gene names | GALNTL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase-like protein 2 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 2) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 2) (Polypeptide GalNAc transferase-like protein 2) (GalNAc-T-like protein 2) (pp-GaNTase-like protein 2). | |||||
|
GLTL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.063779 (rank : 48) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D2N8 | Gene names | Galntl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase-like protein 2 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 2) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 2) (Polypeptide GalNAc transferase-like protein 2) (GalNAc-T-like protein 2) (pp-GaNTase-like protein 2). | |||||
|
GLTL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.060016 (rank : 60) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6IS24, Q8NFV9, Q9NTA8 | Gene names | WBSCR17, GALNTL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 3) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 3) (Polypeptide GalNAc transferase-like protein 3) (GalNAc-T-like protein 3) (pp-GaNTase-like protein 3) (Williams Beuren syndrome chromosome region 17 protein). | |||||
|
GLTL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.059952 (rank : 61) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7TT15, Q8BKN7, Q8BUY1, Q8BX73, Q8BZC8, Q8K483 | Gene names | Wbscr17, Galntl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 3) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 3) (Polypeptide GalNAc transferase-like protein 3) (GalNAc-T-like protein 3) (pp-GaNTase-like protein 3) (Williams-Beuren syndrome chromosome region 17 protein homolog). | |||||
|
GLTL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.059593 (rank : 62) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6P9A2, O95903, Q8NDY9 | Gene names | GALNTL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 4) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 4) (Polypeptide GalNAc transferase-like protein 4) (GalNAc-T-like protein 4) (pp-GaNTase-like protein 4). | |||||
|
GLTL4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.059365 (rank : 63) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K1B9 | Gene names | Galntl4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 4) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 4) (Polypeptide GalNAc transferase-like protein 4) (GalNAc-T-like protein 4) (pp-GaNTase-like protein 4). | |||||
|
GLTL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.062884 (rank : 53) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z4T8, Q75KN2, Q75MD3, Q8NCV4, Q8WW05, Q9UDR9 | Gene names | GALNTL5, GALNT15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 5 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 15) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 15) (Polypeptide GalNAc transferase 15) (GalNAc-T15) (pp-GaNTase 15). | |||||
|
GLTL5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.062682 (rank : 55) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D4M9, Q810S4, Q9CW06 | Gene names | Galntl5, Galnt15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 5 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 15) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 15) (Polypeptide GalNAc transferase 15) (GalNAc-T15) (pp-GaNTase 15). | |||||
|
B4GN4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 129 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GN4_HUMAN
|
||||||
| NC score | 0.962859 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q76KP1, Q96LV2 | Gene names | B4GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
B4GN3_HUMAN
|
||||||
| NC score | 0.931433 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6L9W6, Q6ZNC1, Q8N7T6 | Gene names | B4GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
B4GN3_MOUSE
|
||||||
| NC score | 0.925262 (rank : 4) | θ value | 1.99884e-124 (rank : 4) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6L8S8 | Gene names | B4galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
CGAT1_HUMAN
|
||||||
| NC score | 0.578573 (rank : 5) | θ value | 1.80466e-16 (rank : 5) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TDX6, Q6P9G6, Q8IUF9, Q9NSQ7, Q9NUM9 | Gene names | GALNACT1, CHGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT1_MOUSE
|
||||||
| NC score | 0.575668 (rank : 6) | θ value | 8.95645e-16 (rank : 6) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BJQ9, Q3UZX6, Q8BWV9, Q8BZU7, Q8C195, Q8R0M6 | Gene names | Galnact1, Chgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT2_HUMAN
|
||||||
| NC score | 0.562996 (rank : 7) | θ value | 1.16975e-15 (rank : 7) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8N6G5, Q6MZJ5, Q6MZP6, Q8TCH4, Q9P1I6 | Gene names | GALNACT2, CHGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CGAT2_MOUSE
|
||||||
| NC score | 0.555996 (rank : 8) | θ value | 3.76295e-14 (rank : 8) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C1F4, Q6PAP6, Q8R5A2, Q9D2M1 | Gene names | Galnact2, Chgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CHSS1_HUMAN
|
||||||
| NC score | 0.491343 (rank : 9) | θ value | 9.26847e-13 (rank : 9) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86X52, Q6UX38, Q7LFU5, Q9Y2J5 | Gene names | CHSY1, CHSY, CSS1, KIAA0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase 1) (Chondroitin sulfate synthase 1) (N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS1_MOUSE
|
||||||
| NC score | 0.488152 (rank : 10) | θ value | 6.00763e-12 (rank : 10) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS3_HUMAN
|
||||||
| NC score | 0.470132 (rank : 11) | θ value | 2.13673e-09 (rank : 11) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
CHSS2_HUMAN
|
||||||
| NC score | 0.229030 (rank : 12) | θ value | 0.0563607 (rank : 12) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZ52, Q6UXD6, Q7L4G1, Q9H0F8, Q9H618 | Gene names | CSS2, CHPF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase) (Chondroitin-polymerizing factor). | |||||
|
CHSS2_MOUSE
|
||||||
| NC score | 0.228838 (rank : 13) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6IQX7 | Gene names | Css2, D1Bwg1363e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase). | |||||
|
CHGUT_HUMAN
|
||||||
| NC score | 0.196042 (rank : 14) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2E5, Q6P2I4, Q6UXD2 | Gene names | CSGLCAT, KIAA1402 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate glucuronyltransferase (EC 2.4.1.226) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (Chondroitin glucuronyltransferase II) (CSGlcA-T). | |||||
|
B4GT2_HUMAN
|
||||||
| NC score | 0.110995 (rank : 15) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60909, O60511, Q5T4X5, Q5T4Y5, Q9BUP6, Q9NSY7 | Gene names | B4GALT2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT2_MOUSE
|
||||||
| NC score | 0.109475 (rank : 16) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z2Y2, Q3TMP2 | Gene names | B4galt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
GALT3_MOUSE
|
||||||
| NC score | 0.092676 (rank : 17) | θ value | 0.125558 (rank : 17) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70419, Q80V55 | Gene names | Galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 3) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 3) (Polypeptide GalNAc transferase 3) (GalNAc-T3) (pp-GaNTase 3). | |||||
|
B4GT6_HUMAN
|
||||||
| NC score | 0.092463 (rank : 18) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UBX8, O60514 | Gene names | B4GALT6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
B4GT6_MOUSE
|
||||||
| NC score | 0.092022 (rank : 19) | θ value | 0.125558 (rank : 16) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WVK5 | Gene names | B4galt6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
GALT3_HUMAN
|
||||||
| NC score | 0.090650 (rank : 20) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14435, Q7Z476 | Gene names | GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 3) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 3) (Polypeptide GalNAc transferase 3) (GalNAc-T3) (pp-GaNTase 3). | |||||
|
GALT6_HUMAN
|
||||||
| NC score | 0.089674 (rank : 21) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NCL4, Q8IYH4, Q9H6G2, Q9UIV5 | Gene names | GALNT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
B4GT5_MOUSE
|
||||||
| NC score | 0.086699 (rank : 22) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JMK0 | Gene names | B4galt5, Bgt-5 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
B4GT5_HUMAN
|
||||||
| NC score | 0.086222 (rank : 23) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43286, Q2M394, Q9UJQ8 | Gene names | B4GALT5 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 5 (EC 2.4.1.-) (Beta-1,4-GalTase 5) (Beta4Gal-T5) (b4Gal-T5) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 5) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 5) (Beta-1,4-GalT II). | |||||
|
GALT6_MOUSE
|
||||||
| NC score | 0.084218 (rank : 24) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8C7U7, Q3TWF0, Q8CED2, Q9QZ16 | Gene names | Galnt6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
GLT12_MOUSE
|
||||||
| NC score | 0.077712 (rank : 25) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BGT9 | Gene names | Galnt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 12 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 12) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 12) (Polypeptide GalNAc transferase 12) (GalNAc-T12) (pp-GaNTase 12). | |||||
|
GALT4_HUMAN
|
||||||
| NC score | 0.076817 (rank : 26) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N4A0, O00208 | Gene names | GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 4) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 4) (Polypeptide GalNAc transferase 4) (GalNAc-T4) (pp-GaNTase 4). | |||||
|
GALT4_MOUSE
|
||||||
| NC score | 0.076806 (rank : 27) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O08832, Q3U681 | Gene names | Galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 4) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 4) (Polypeptide GalNAc transferase 4) (GalNAc-T4) (pp-GaNTase 4). | |||||
|
GLT14_HUMAN
|
||||||
| NC score | 0.075908 (rank : 28) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96FL9, Q8IVI4, Q9BRH1, Q9H827, Q9H9J8 | Gene names | GALNT14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 14 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 14) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 14) (Polypeptide GalNAc transferase 14) (GalNAc-T14) (pp-GaNTase 14). | |||||
|
GLT14_MOUSE
|
||||||
| NC score | 0.075869 (rank : 29) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BVG5, Q60GT2, Q8BTI6, Q9DCJ2 | Gene names | Galnt14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 14 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 14) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 14) (Polypeptide GalNAc transferase 14) (GalNAc-T14) (pp-GaNTase 14). | |||||
|
GLTL1_MOUSE
|
||||||
| NC score | 0.075844 (rank : 30) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JJ61 | Gene names | Galntl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 1 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 1) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 1) (Polypeptide GalNAc transferase-like protein 1) (GalNAc-T-like protein 1) (pp-GaNTase-like protein 1). | |||||
|
GLTL1_HUMAN
|
||||||
| NC score | 0.075825 (rank : 31) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N428, Q9ULT9 | Gene names | GALNTL1, KIAA1130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 1 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 1) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 1) (Polypeptide GalNAc transferase-like protein 1) (GalNAc-T-like protein 1) (pp-GaNTase-like protein 1). | |||||
|
GLT13_MOUSE
|
||||||
| NC score | 0.075492 (rank : 32) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CF93, Q8BLE4, Q8BYT3 | Gene names | Galnt13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 13 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 13) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 13) (Polypeptide GalNAc transferase 13) (GalNAc-T13) (pp-GaNTase 13). | |||||
|
GLT13_HUMAN
|
||||||
| NC score | 0.075481 (rank : 33) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IUC8, Q6ZWG1, Q96PX0, Q9UIE5 | Gene names | GALNT13, KIAA1918 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 13 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 13) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 13) (Polypeptide GalNAc transferase 13) (GalNAc-T13) (pp-GaNTase 13). | |||||
|
GALT5_MOUSE
|
||||||
| NC score | 0.075024 (rank : 34) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8C102 | Gene names | Galnt5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
GLT12_HUMAN
|
||||||
| NC score | 0.074664 (rank : 35) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IXK2, Q5TCF7, Q8NG54, Q96CT9, Q9H771 | Gene names | GALNT12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 12 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 12) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 12) (Polypeptide GalNAc transferase 12) (GalNAc-T12) (pp-GaNTase 12). | |||||
|
GALT5_HUMAN
|
||||||
| NC score | 0.073879 (rank : 36) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z7M9, Q9UGK7, Q9UHL6 | Gene names | GALNT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
GALT1_HUMAN
|
||||||
| NC score | 0.071039 (rank : 37) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q10472, Q86TJ7, Q9UM86 | Gene names | GALNT1 | |||
|
Domain Architecture |

|
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 1 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 1) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 1) (Polypeptide GalNAc transferase 1) (GalNAc-T1) (pp-GaNTase 1) [Contains: Polypeptide N-acetylgalactosaminyltransferase 1 soluble form]. | |||||
|
GALT1_MOUSE
|
||||||
| NC score | 0.071009 (rank : 38) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08912, Q5BKS3, Q7TND1 | Gene names | Galnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 1 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 1) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 1) (Polypeptide GalNAc transferase 1) (GalNAc-T1) (pp-GaNTase 1) [Contains: Polypeptide N-acetylgalactosaminyltransferase 1 soluble form]. | |||||
|
GALT2_HUMAN
|
||||||
| NC score | 0.070121 (rank : 39) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q10471, Q9NPY4 | Gene names | GALNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 2 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 2) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 2) (Polypeptide GalNAc transferase 2) (GalNAc-T2) (pp-GaNTase 2) [Contains: Polypeptide N-acetylgalactosaminyltransferase 2 soluble form]. | |||||
|
GALT2_MOUSE
|
||||||
| NC score | 0.069917 (rank : 40) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PB93, Q7TSI5, Q8BL27, Q922K5, Q99ME1 | Gene names | Galnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 2 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 2) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 2) (Polypeptide GalNAc transferase 2) (GalNAc-T2) (pp-GaNTase 2) [Contains: Polypeptide N-acetylgalactosaminyltransferase 2 soluble form]. | |||||
|
WASP_MOUSE
|
||||||
| NC score | 0.069442 (rank : 41) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
WASP_HUMAN
|
||||||
| NC score | 0.067740 (rank : 42) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
B4GT1_MOUSE
|
||||||
| NC score | 0.067415 (rank : 43) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P15535 | Gene names | B4galt1, Ggtb, Ggtb2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
GLT10_HUMAN
|
||||||
| NC score | 0.066929 (rank : 44) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86SR1, Q6IN56, Q86VP8, Q8IXJ2, Q8TEJ2, Q96IV2, Q9H8E1, Q9Y4M4 | Gene names | GALNT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 10 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 10) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 10) (Polypeptide GalNAc transferase 10) (GalNAc-T10) (pp-GaNTase 10). | |||||
|
GLT10_MOUSE
|
||||||
| NC score | 0.066885 (rank : 45) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6P9S7, Q6KAQ2, Q8BZU8, Q91YJ6 | Gene names | Galnt10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 10 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 10) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 10) (Polypeptide GalNAc transferase 10) (GalNAc-T10) (pp-GaNTase 10). | |||||
|
GLTL2_HUMAN
|
||||||
| NC score | 0.063976 (rank : 46) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N3T1, Q86T60, Q96C46, Q96DJ5 | Gene names | GALNTL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase-like protein 2 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 2) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 2) (Polypeptide GalNAc transferase-like protein 2) (GalNAc-T-like protein 2) (pp-GaNTase-like protein 2). | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.063915 (rank : 47) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
GLTL2_MOUSE
|
||||||
| NC score | 0.063779 (rank : 48) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D2N8 | Gene names | Galntl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase-like protein 2 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 2) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 2) (Polypeptide GalNAc transferase-like protein 2) (GalNAc-T-like protein 2) (pp-GaNTase-like protein 2). | |||||
|
GLT11_HUMAN
|
||||||
| NC score | 0.063271 (rank : 49) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NCW6, Q6PCD1, Q9H6C2, Q9H6Z5, Q9UDR8 | Gene names | GALNT11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 11 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 11) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 11) (Polypeptide GalNAc transferase 11) (GalNAc-T11) (pp-GaNTase 11). | |||||
|
GLT11_MOUSE
|
||||||
| NC score | 0.063224 (rank : 50) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q921L8, Q3TT83, Q8BU26, Q8K032 | Gene names | Galnt11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 11 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 11) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 11) (Polypeptide GalNAc transferase 11) (GalNAc-T11) (pp-GaNTase 11). | |||||
|
GALT7_MOUSE
|
||||||
| NC score | 0.062962 (rank : 51) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q80VA0, Q8BY62, Q8BZ70, Q8BZW0, Q91VW6, Q99MD7 | Gene names | Galnt7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylgalactosaminyltransferase 7 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 7) (UDP-GalNAc:polypeptide N- acetylgalactosaminyltransferase 7) (Polypeptide GalNAc transferase 7) (GalNAc-T7) (pp-GaNTase 7). | |||||
|
B4GT1_HUMAN
|
||||||
| NC score | 0.062922 (rank : 52) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
GLTL5_HUMAN
|
||||||
| NC score | 0.062884 (rank : 53) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z4T8, Q75KN2, Q75MD3, Q8NCV4, Q8WW05, Q9UDR9 | Gene names | GALNTL5, GALNT15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 5 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 15) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 15) (Polypeptide GalNAc transferase 15) (GalNAc-T15) (pp-GaNTase 15). | |||||
|
GALT7_HUMAN
|
||||||
| NC score | 0.062788 (rank : 54) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86SF2, Q7Z5W7, Q9UJ28 | Gene names | GALNT7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylgalactosaminyltransferase 7 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 7) (UDP-GalNAc:polypeptide N- acetylgalactosaminyltransferase 7) (Polypeptide GalNAc transferase 7) (GalNAc-T7) (pp-GaNTase 7). | |||||
|
GLTL5_MOUSE
|
||||||
| NC score | 0.062682 (rank : 55) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D4M9, Q810S4, Q9CW06 | Gene names | Galntl5, Galnt15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 5 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 15) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 15) (Polypeptide GalNAc transferase 15) (GalNAc-T15) (pp-GaNTase 15). | |||||
|
B4GT4_MOUSE
|
||||||
| NC score | 0.061981 (rank : 56) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JJ04, Q8BR54, Q9QY12 | Gene names | B4galt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
GALT9_HUMAN
|
||||||
| NC score | 0.060501 (rank : 57) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HCQ5, Q6NT54, Q8NFR1 | Gene names | GALNT9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 9 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 9) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 9) (Polypeptide GalNAc transferase 9) (GalNAc-T9) (pp-GaNTase 9). | |||||
|
GALT8_HUMAN
|
||||||
| NC score | 0.060299 (rank : 58) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NY28 | Gene names | GALNT8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable polypeptide N-acetylgalactosaminyltransferase 8 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 8) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 8) (Polypeptide GalNAc transferase 8) (GalNAc-T8) (pp-GaNTase 8). | |||||
|
B4GT4_HUMAN
|
||||||
| NC score | 0.060232 (rank : 59) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60513, Q68D68, Q9BSW3, Q9C078 | Gene names | B4GALT4 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 4 (EC 2.4.1.-) (Beta-1,4-GalTase 4) (Beta4Gal-T4) (b4Gal-T4) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 4) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 4) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
GLTL3_HUMAN
|
||||||
| NC score | 0.060016 (rank : 60) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6IS24, Q8NFV9, Q9NTA8 | Gene names | WBSCR17, GALNTL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 3) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 3) (Polypeptide GalNAc transferase-like protein 3) (GalNAc-T-like protein 3) (pp-GaNTase-like protein 3) (Williams Beuren syndrome chromosome region 17 protein). | |||||
|
GLTL3_MOUSE
|
||||||
| NC score | 0.059952 (rank : 61) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7TT15, Q8BKN7, Q8BUY1, Q8BX73, Q8BZC8, Q8K483 | Gene names | Wbscr17, Galntl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 3) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 3) (Polypeptide GalNAc transferase-like protein 3) (GalNAc-T-like protein 3) (pp-GaNTase-like protein 3) (Williams-Beuren syndrome chromosome region 17 protein homolog). | |||||
|
GLTL4_HUMAN
|
||||||
| NC score | 0.059593 (rank : 62) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6P9A2, O95903, Q8NDY9 | Gene names | GALNTL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 4) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 4) (Polypeptide GalNAc transferase-like protein 4) (GalNAc-T-like protein 4) (pp-GaNTase-like protein 4). | |||||
|
GLTL4_MOUSE
|
||||||
| NC score | 0.059365 (rank : 63) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K1B9 | Gene names | Galntl4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 4) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 4) (Polypeptide GalNAc transferase-like protein 4) (GalNAc-T-like protein 4) (pp-GaNTase-like protein 4). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.055822 (rank : 64) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
B3GLT_MOUSE
|
||||||
| NC score | 0.055562 (rank : 65) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BHT6, Q0PCR7, Q149S5 | Gene names | B3galtl, Gm1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.054776 (rank : 66) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
B4GT3_MOUSE
|
||||||
| NC score | 0.053814 (rank : 67) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91YY2, Q9QY13 | Gene names | B4galt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B4GT3_HUMAN
|
||||||
| NC score | 0.053495 (rank : 68) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60512, O60910, Q9BPZ4, Q9H8T2 | Gene names | B4GALT3 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 3 (EC 2.4.1.-) (Beta-1,4-GalTase 3) (Beta4Gal-T3) (b4Gal-T3) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 3) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 3) [Includes: N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase (EC 2.4.1.38); Beta-N- acetylglucosaminyl-glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
B3GLT_HUMAN
|
||||||
| NC score | 0.051502 (rank : 69) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6Y288, Q5W0H2, Q6NUI3 | Gene names | B3GALTL, B3GTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.050830 (rank : 70) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.046006 (rank : 71) | θ value | 0.0961366 (rank : 14) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.044531 (rank : 72) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.042300 (rank : 73) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.040725 (rank : 74) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.040600 (rank : 75) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.038872 (rank : 76) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
GA2L2_MOUSE
|
||||||
| NC score | 0.038373 (rank : 77) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
RUSC1_MOUSE
|
||||||
| NC score | 0.037270 (rank : 78) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BG26, Q6PHT9 | Gene names | Rusc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.036196 (rank : 79) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
3BP2_HUMAN
|
||||||
| NC score | 0.036046 (rank : 80) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
EFS_HUMAN
|
||||||
| NC score | 0.036037 (rank : 81) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
CSPG5_MOUSE
|
||||||
| NC score | 0.034758 (rank : 82) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q71M36, Q71M37, Q7TNT8, Q8BPJ5, Q9QY32 | Gene names | Cspg5, Caleb, Ngc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.033665 (rank : 83) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
LEUK_MOUSE
|
||||||
| NC score | 0.033344 (rank : 84) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15702 | Gene names | Spn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Ly- 48) (B cell differentiation antigen LP-3) (CD43 antigen). | |||||
|
LR37B_HUMAN
|
||||||
| NC score | 0.031720 (rank : 85) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.030211 (rank : 86) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
T240L_HUMAN
|
||||||
| NC score | 0.029540 (rank : 87) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q71F56, Q68DN4, Q9H8C0, Q9NSY9, Q9UFD8, Q9UPX5 | Gene names | THRAP2, KIAA1025, PROSIT240, TRAP240L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.028393 (rank : 88) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.028383 (rank : 89) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
K1949_HUMAN
|
||||||
| NC score | 0.026842 (rank : 90) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6NYC8, Q68CK8, Q6ZUJ6, Q8NDQ4, Q8TF52, Q9BRL9 | Gene names | KIAA1949 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.026378 (rank : 91) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.026336 (rank : 92) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
FMNL_HUMAN
|
||||||
| NC score | 0.024922 (rank : 93) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
FMN2_MOUSE
|
||||||
| NC score | 0.023113 (rank : 94) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.022761 (rank : 95) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.022374 (rank : 96) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
S2A4R_HUMAN
|
||||||
| NC score | 0.022261 (rank : 97) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NR83, Q6F6I6, Q6F6I7, Q6GTK5, Q8TAH5, Q8WVW7, Q96QD3, Q9BV85 | Gene names | SLC2A4RG, HDBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SLC2A4 regulator (GLUT4 enhancer factor) (GEF) (Huntington disease gene regulatory region-binding protein 1) (HDBP-1). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.021951 (rank : 98) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
TM108_MOUSE
|
||||||
| NC score | 0.021823 (rank : 99) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
CPSF6_MOUSE
|
||||||
| NC score | 0.021218 (rank : 100) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
CPSF6_HUMAN
|
||||||
| NC score | 0.021117 (rank : 101) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
BSCL2_HUMAN
|
||||||
| NC score | 0.020865 (rank : 102) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
SFR15_HUMAN
|
||||||
| NC score | 0.020667 (rank : 103) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
FMNL_MOUSE
|
||||||
| NC score | 0.020135 (rank : 104) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JL26, Q6KAN4, Q9Z2V7 | Gene names | Fmnl1, Frl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-like protein 1 (Formin-related protein). | |||||
|
DIAP1_MOUSE
|
||||||
| NC score | 0.020113 (rank : 105) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
SSH3_HUMAN
|
||||||
| NC score | 0.019490 (rank : 106) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TE77, Q6PK42, Q76I75, Q8N9L8, Q8WYL0, Q9NV45, Q9NWZ7 | Gene names | SSH3, SSH3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (hSSH-3L). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.018894 (rank : 107) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.018169 (rank : 108) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.018052 (rank : 109) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
TPP1_HUMAN
|
||||||
| NC score | 0.017998 (rank : 110) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14773, Q53HT1, Q5JAK6, Q6UX56, Q96C37 | Gene names | TPP1, CLN2, GIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Tripeptidyl-peptidase 1 precursor (EC 3.4.14.9) (Tripeptidyl-peptidase I) (TPP-I) (Tripeptidyl aminopeptidase) (Lysosomal pepstatin insensitive protease) (LPIC) (Growth-inhibiting gene 1 protein). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.017993 (rank : 111) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.016282 (rank : 112) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
FGD1_HUMAN
|
||||||
| NC score | 0.016015 (rank : 113) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
LEG3_MOUSE
|
||||||
| NC score | 0.015946 (rank : 114) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P16110 | Gene names | Lgals3 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-34 galactoside-binding lectin). | |||||
|
WASF2_HUMAN
|
||||||
| NC score | 0.015756 (rank : 115) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.015723 (rank : 116) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
RHG07_MOUSE
|
||||||
| NC score | 0.015586 (rank : 117) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
TMUB2_MOUSE
|
||||||
| NC score | 0.015120 (rank : 118) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3V209, Q8K0X0, Q91YQ0, Q9D8C5 | Gene names | Tmub2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 2. | |||||
|
FGD1_MOUSE
|
||||||
| NC score | 0.014877 (rank : 119) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.014283 (rank : 120) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
PDE1C_HUMAN
|
||||||
| NC score | 0.014098 (rank : 121) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.013831 (rank : 122) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.013641 (rank : 123) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
COIA1_HUMAN
|
||||||
| NC score | 0.013062 (rank : 124) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
DYN3_HUMAN
|
||||||
| NC score | 0.012873 (rank : 125) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
LTBP3_HUMAN
|
||||||
| NC score | 0.012799 (rank : 126) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NS15, O15107, Q96HB9, Q9H7K2, Q9UFN4 | Gene names | LTBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
TNFL6_HUMAN
|
||||||
| NC score | 0.012286 (rank : 127) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48023, Q9BZP9 | Gene names | FASLG, APT1LG1, FASL, TNFSF6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor ligand superfamily member 6 (Fas antigen ligand) (Fas ligand) (CD178 antigen) (CD95L protein) (Apoptosis antigen ligand) (APTL) [Contains: Tumor necrosis factor ligand superfamily member 6, membrane form; Tumor necrosis factor ligand superfamily member 6, soluble form]. | |||||
|
PCX1_HUMAN
|
||||||
| NC score | 0.012177 (rank : 128) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96RV3, O94897, Q96AI7, Q9Y2J9 | Gene names | PCNX, KIAA0805, KIAA0995, PCNXL1 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 1 (Pecanex homolog). | |||||
|
IF32_HUMAN
|
||||||
| NC score | 0.011773 (rank : 129) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13347 | Gene names | EIF3S2, TRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 2 (eIF-3 beta) (eIF3 p36) (eIF3i) (TGF-beta receptor-interacting protein 1) (TRIP-1). | |||||
|
IF32_MOUSE
|
||||||
| NC score | 0.011771 (rank : 130) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZD9 | Gene names | Eif3s2, Trip1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 2 (eIF-3 beta) (eIF3 p36) (eIF3i) (TGF-beta receptor-interacting protein 1) (TRIP-1). | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.010047 (rank : 131) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.009827 (rank : 132) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.009430 (rank : 133) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
IASPP_HUMAN
|
||||||
| NC score | 0.009233 (rank : 134) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
SC6A5_MOUSE
|
||||||
| NC score | 0.008699 (rank : 135) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q761V0, Q8CFM5, Q91ZQ2 | Gene names | Slc6a5, Glyt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
MICA3_HUMAN
|
||||||
| NC score | 0.008480 (rank : 136) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7RTP6, Q6ICK4, Q9P2I3 | Gene names | MICAL3, KIAA1364 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-3. | |||||
|
NEBL_HUMAN
|
||||||
| NC score | 0.007996 (rank : 137) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O76041, Q9UIC4 | Gene names | NEBL | |||
|
Domain Architecture |

|
|||||
| Description | Nebulette (Actin-binding Z-disk protein). | |||||
|
ETV3_MOUSE
|
||||||
| NC score | 0.006730 (rank : 138) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
LIPB2_MOUSE
|
||||||
| NC score | 0.006322 (rank : 139) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35711, Q7TMG4, Q8CBS6, Q99KX6 | Gene names | Ppfibp2, Cclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2) (Coiled-coil-like protein 1). | |||||
|
RTN4R_HUMAN
|
||||||
| NC score | 0.006141 (rank : 140) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BZR6 | Gene names | RTN4R, NOGOR | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 receptor precursor (Nogo receptor) (NgR) (Nogo-66 receptor). | |||||
|
JHD3B_MOUSE
|
||||||
| NC score | 0.005017 (rank : 141) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.004695 (rank : 142) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
POGZ_MOUSE
|
||||||
| NC score | 0.003948 (rank : 143) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BZH4, Q4VA94, Q80TZ8, Q8C0K1, Q8K294 | Gene names | Pogz, Kiaa0461 | |||
|
Domain Architecture |

|
|||||
| Description | Pogo transposable element with ZNF domain. | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.003672 (rank : 144) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
LATS1_MOUSE
|
||||||
| NC score | 0.002960 (rank : 145) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
ESR1_HUMAN
|
||||||
| NC score | 0.002810 (rank : 146) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P03372, Q13511, Q14276, Q9NU51, Q9UDZ7, Q9UIS7 | Gene names | ESR1, ESR, NR3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen receptor (ER) (Estradiol receptor) (ER-alpha). | |||||
|
FOG1_HUMAN
|
||||||
| NC score | 0.002716 (rank : 147) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
LATS1_HUMAN
|
||||||
| NC score | 0.002691 (rank : 148) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95835, Q6PKD0 | Gene names | LATS1, WARTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase) (h-warts). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.002538 (rank : 149) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
KIF3C_HUMAN
|
||||||
| NC score | 0.002411 (rank : 150) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14782, O43544 | Gene names | KIF3C | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
PAK7_MOUSE
|
||||||
| NC score | 0.000238 (rank : 151) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 129 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C015, Q3TQJ7, Q6RWS7 | Gene names | Pak7, Pak5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PAK 7 (EC 2.7.11.1) (p21-activated kinase 7) (PAK-7) (p21-activated kinase 5). | |||||