Please be patient as the page loads
|
GALT8_HUMAN
|
||||||
| SwissProt Accessions | Q9NY28 | Gene names | GALNT8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable polypeptide N-acetylgalactosaminyltransferase 8 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 8) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 8) (Polypeptide GalNAc transferase 8) (GalNAc-T8) (pp-GaNTase 8). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GALT8_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9NY28 | Gene names | GALNT8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable polypeptide N-acetylgalactosaminyltransferase 8 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 8) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 8) (Polypeptide GalNAc transferase 8) (GalNAc-T8) (pp-GaNTase 8). | |||||
|
GLTL4_HUMAN
|
||||||
| θ value | 5.94893e-161 (rank : 2) | NC score | 0.993910 (rank : 2) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6P9A2, O95903, Q8NDY9 | Gene names | GALNTL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 4) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 4) (Polypeptide GalNAc transferase-like protein 4) (GalNAc-T-like protein 4) (pp-GaNTase-like protein 4). | |||||
|
GLTL4_MOUSE
|
||||||
| θ value | 6.1562e-158 (rank : 3) | NC score | 0.993593 (rank : 4) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K1B9 | Gene names | Galntl4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 4) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 4) (Polypeptide GalNAc transferase-like protein 4) (GalNAc-T-like protein 4) (pp-GaNTase-like protein 4). | |||||
|
GALT9_HUMAN
|
||||||
| θ value | 4.73554e-142 (rank : 4) | NC score | 0.993803 (rank : 3) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HCQ5, Q6NT54, Q8NFR1 | Gene names | GALNT9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 9 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 9) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 9) (Polypeptide GalNAc transferase 9) (GalNAc-T9) (pp-GaNTase 9). | |||||
|
GLTL3_HUMAN
|
||||||
| θ value | 7.56041e-140 (rank : 5) | NC score | 0.992529 (rank : 5) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6IS24, Q8NFV9, Q9NTA8 | Gene names | WBSCR17, GALNTL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 3) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 3) (Polypeptide GalNAc transferase-like protein 3) (GalNAc-T-like protein 3) (pp-GaNTase-like protein 3) (Williams Beuren syndrome chromosome region 17 protein). | |||||
|
GLTL3_MOUSE
|
||||||
| θ value | 2.19974e-139 (rank : 6) | NC score | 0.992451 (rank : 6) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TT15, Q8BKN7, Q8BUY1, Q8BX73, Q8BZC8, Q8K483 | Gene names | Wbscr17, Galntl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 3) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 3) (Polypeptide GalNAc transferase-like protein 3) (GalNAc-T-like protein 3) (pp-GaNTase-like protein 3) (Williams-Beuren syndrome chromosome region 17 protein homolog). | |||||
|
GLT13_HUMAN
|
||||||
| θ value | 2.31364e-80 (rank : 7) | NC score | 0.975513 (rank : 13) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IUC8, Q6ZWG1, Q96PX0, Q9UIE5 | Gene names | GALNT13, KIAA1918 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 13 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 13) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 13) (Polypeptide GalNAc transferase 13) (GalNAc-T13) (pp-GaNTase 13). | |||||
|
GLT11_HUMAN
|
||||||
| θ value | 3.0217e-80 (rank : 8) | NC score | 0.977464 (rank : 8) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8NCW6, Q6PCD1, Q9H6C2, Q9H6Z5, Q9UDR8 | Gene names | GALNT11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 11 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 11) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 11) (Polypeptide GalNAc transferase 11) (GalNAc-T11) (pp-GaNTase 11). | |||||
|
GLT13_MOUSE
|
||||||
| θ value | 3.94648e-80 (rank : 9) | NC score | 0.975509 (rank : 14) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CF93, Q8BLE4, Q8BYT3 | Gene names | Galnt13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 13 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 13) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 13) (Polypeptide GalNAc transferase 13) (GalNAc-T13) (pp-GaNTase 13). | |||||
|
GALT1_HUMAN
|
||||||
| θ value | 2.55803e-79 (rank : 10) | NC score | 0.975527 (rank : 12) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q10472, Q86TJ7, Q9UM86 | Gene names | GALNT1 | |||
|
Domain Architecture |

|
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 1 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 1) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 1) (Polypeptide GalNAc transferase 1) (GalNAc-T1) (pp-GaNTase 1) [Contains: Polypeptide N-acetylgalactosaminyltransferase 1 soluble form]. | |||||
|
GLT11_MOUSE
|
||||||
| θ value | 4.36332e-79 (rank : 11) | NC score | 0.977406 (rank : 10) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q921L8, Q3TT83, Q8BU26, Q8K032 | Gene names | Galnt11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 11 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 11) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 11) (Polypeptide GalNAc transferase 11) (GalNAc-T11) (pp-GaNTase 11). | |||||
|
GALT1_MOUSE
|
||||||
| θ value | 5.69867e-79 (rank : 12) | NC score | 0.975826 (rank : 11) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O08912, Q5BKS3, Q7TND1 | Gene names | Galnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 1 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 1) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 1) (Polypeptide GalNAc transferase 1) (GalNAc-T1) (pp-GaNTase 1) [Contains: Polypeptide N-acetylgalactosaminyltransferase 1 soluble form]. | |||||
|
GALT4_MOUSE
|
||||||
| θ value | 7.44272e-79 (rank : 13) | NC score | 0.974317 (rank : 15) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O08832, Q3U681 | Gene names | Galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 4) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 4) (Polypeptide GalNAc transferase 4) (GalNAc-T4) (pp-GaNTase 4). | |||||
|
GALT4_HUMAN
|
||||||
| θ value | 2.02684e-76 (rank : 14) | NC score | 0.974237 (rank : 16) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N4A0, O00208 | Gene names | GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 4) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 4) (Polypeptide GalNAc transferase 4) (GalNAc-T4) (pp-GaNTase 4). | |||||
|
GALT6_MOUSE
|
||||||
| θ value | 4.67263e-73 (rank : 15) | NC score | 0.970849 (rank : 27) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8C7U7, Q3TWF0, Q8CED2, Q9QZ16 | Gene names | Galnt6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
GALT6_HUMAN
|
||||||
| θ value | 1.04096e-72 (rank : 16) | NC score | 0.970743 (rank : 29) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NCL4, Q8IYH4, Q9H6G2, Q9UIV5 | Gene names | GALNT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
GLTL5_MOUSE
|
||||||
| θ value | 3.34864e-71 (rank : 17) | NC score | 0.979839 (rank : 7) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D4M9, Q810S4, Q9CW06 | Gene names | Galntl5, Galnt15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 5 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 15) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 15) (Polypeptide GalNAc transferase 15) (GalNAc-T15) (pp-GaNTase 15). | |||||
|
GALT2_HUMAN
|
||||||
| θ value | 5.71191e-71 (rank : 18) | NC score | 0.973911 (rank : 18) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q10471, Q9NPY4 | Gene names | GALNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 2 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 2) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 2) (Polypeptide GalNAc transferase 2) (GalNAc-T2) (pp-GaNTase 2) [Contains: Polypeptide N-acetylgalactosaminyltransferase 2 soluble form]. | |||||
|
GALT2_MOUSE
|
||||||
| θ value | 5.71191e-71 (rank : 19) | NC score | 0.973926 (rank : 17) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6PB93, Q7TSI5, Q8BL27, Q922K5, Q99ME1 | Gene names | Galnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 2 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 2) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 2) (Polypeptide GalNAc transferase 2) (GalNAc-T2) (pp-GaNTase 2) [Contains: Polypeptide N-acetylgalactosaminyltransferase 2 soluble form]. | |||||
|
GLTL1_HUMAN
|
||||||
| θ value | 8.24801e-70 (rank : 20) | NC score | 0.972312 (rank : 23) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N428, Q9ULT9 | Gene names | GALNTL1, KIAA1130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 1 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 1) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 1) (Polypeptide GalNAc transferase-like protein 1) (GalNAc-T-like protein 1) (pp-GaNTase-like protein 1). | |||||
|
GALT3_HUMAN
|
||||||
| θ value | 1.40689e-69 (rank : 21) | NC score | 0.969192 (rank : 34) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q14435, Q7Z476 | Gene names | GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 3) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 3) (Polypeptide GalNAc transferase 3) (GalNAc-T3) (pp-GaNTase 3). | |||||
|
GALT5_HUMAN
|
||||||
| θ value | 1.40689e-69 (rank : 22) | NC score | 0.968463 (rank : 36) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z7M9, Q9UGK7, Q9UHL6 | Gene names | GALNT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
GLTL1_MOUSE
|
||||||
| θ value | 4.09345e-69 (rank : 23) | NC score | 0.972284 (rank : 24) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JJ61 | Gene names | Galntl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 1 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 1) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 1) (Polypeptide GalNAc transferase-like protein 1) (GalNAc-T-like protein 1) (pp-GaNTase-like protein 1). | |||||
|
GALT5_MOUSE
|
||||||
| θ value | 5.34618e-69 (rank : 24) | NC score | 0.970583 (rank : 30) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8C102 | Gene names | Galnt5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
GALT3_MOUSE
|
||||||
| θ value | 6.98233e-69 (rank : 25) | NC score | 0.969456 (rank : 33) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70419, Q80V55 | Gene names | Galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 3) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 3) (Polypeptide GalNAc transferase 3) (GalNAc-T3) (pp-GaNTase 3). | |||||
|
GLT14_HUMAN
|
||||||
| θ value | 1.11475e-66 (rank : 26) | NC score | 0.971398 (rank : 26) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96FL9, Q8IVI4, Q9BRH1, Q9H827, Q9H9J8 | Gene names | GALNT14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 14 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 14) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 14) (Polypeptide GalNAc transferase 14) (GalNAc-T14) (pp-GaNTase 14). | |||||
|
GLT12_HUMAN
|
||||||
| θ value | 4.23606e-66 (rank : 27) | NC score | 0.973648 (rank : 20) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IXK2, Q5TCF7, Q8NG54, Q96CT9, Q9H771 | Gene names | GALNT12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 12 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 12) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 12) (Polypeptide GalNAc transferase 12) (GalNAc-T12) (pp-GaNTase 12). | |||||
|
GLT10_MOUSE
|
||||||
| θ value | 9.43692e-66 (rank : 28) | NC score | 0.973548 (rank : 21) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6P9S7, Q6KAQ2, Q8BZU8, Q91YJ6 | Gene names | Galnt10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 10 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 10) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 10) (Polypeptide GalNAc transferase 10) (GalNAc-T10) (pp-GaNTase 10). | |||||
|
GLTL2_HUMAN
|
||||||
| θ value | 9.43692e-66 (rank : 29) | NC score | 0.972149 (rank : 25) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N3T1, Q86T60, Q96C46, Q96DJ5 | Gene names | GALNTL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase-like protein 2 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 2) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 2) (Polypeptide GalNAc transferase-like protein 2) (GalNAc-T-like protein 2) (pp-GaNTase-like protein 2). | |||||
|
GLTL5_HUMAN
|
||||||
| θ value | 1.2325e-65 (rank : 30) | NC score | 0.977455 (rank : 9) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z4T8, Q75KN2, Q75MD3, Q8NCV4, Q8WW05, Q9UDR9 | Gene names | GALNTL5, GALNT15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 5 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 15) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 15) (Polypeptide GalNAc transferase 15) (GalNAc-T15) (pp-GaNTase 15). | |||||
|
GLT10_HUMAN
|
||||||
| θ value | 1.6097e-65 (rank : 31) | NC score | 0.973441 (rank : 22) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q86SR1, Q6IN56, Q86VP8, Q8IXJ2, Q8TEJ2, Q96IV2, Q9H8E1, Q9Y4M4 | Gene names | GALNT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 10 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 10) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 10) (Polypeptide GalNAc transferase 10) (GalNAc-T10) (pp-GaNTase 10). | |||||
|
GLT12_MOUSE
|
||||||
| θ value | 7.98882e-65 (rank : 32) | NC score | 0.973723 (rank : 19) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BGT9 | Gene names | Galnt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 12 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 12) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 12) (Polypeptide GalNAc transferase 12) (GalNAc-T12) (pp-GaNTase 12). | |||||
|
GLT14_MOUSE
|
||||||
| θ value | 1.04338e-64 (rank : 33) | NC score | 0.970793 (rank : 28) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BVG5, Q60GT2, Q8BTI6, Q9DCJ2 | Gene names | Galnt14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 14 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 14) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 14) (Polypeptide GalNAc transferase 14) (GalNAc-T14) (pp-GaNTase 14). | |||||
|
GALT7_HUMAN
|
||||||
| θ value | 2.56992e-63 (rank : 34) | NC score | 0.970319 (rank : 31) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q86SF2, Q7Z5W7, Q9UJ28 | Gene names | GALNT7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylgalactosaminyltransferase 7 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 7) (UDP-GalNAc:polypeptide N- acetylgalactosaminyltransferase 7) (Polypeptide GalNAc transferase 7) (GalNAc-T7) (pp-GaNTase 7). | |||||
|
GALT7_MOUSE
|
||||||
| θ value | 1.66577e-62 (rank : 35) | NC score | 0.970160 (rank : 32) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80VA0, Q8BY62, Q8BZ70, Q8BZW0, Q91VW6, Q99MD7 | Gene names | Galnt7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylgalactosaminyltransferase 7 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 7) (UDP-GalNAc:polypeptide N- acetylgalactosaminyltransferase 7) (Polypeptide GalNAc transferase 7) (GalNAc-T7) (pp-GaNTase 7). | |||||
|
GLTL2_MOUSE
|
||||||
| θ value | 3.974e-56 (rank : 36) | NC score | 0.968998 (rank : 35) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D2N8 | Gene names | Galntl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase-like protein 2 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 2) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 2) (Polypeptide GalNAc transferase-like protein 2) (GalNAc-T-like protein 2) (pp-GaNTase-like protein 2). | |||||
|
CE152_HUMAN
|
||||||
| θ value | 0.163984 (rank : 37) | NC score | 0.019123 (rank : 42) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 0.279714 (rank : 38) | NC score | 0.014593 (rank : 44) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
RAI14_HUMAN
|
||||||
| θ value | 0.365318 (rank : 39) | NC score | 0.007448 (rank : 76) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.014831 (rank : 43) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
CT096_HUMAN
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.023315 (rank : 40) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NUD7, Q8N840, Q8NAX5 | Gene names | C20orf96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf96. | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.011468 (rank : 60) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
CA049_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.019160 (rank : 41) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5T0J7, Q5T0J9, Q8IYU6, Q8N7Z1, Q8NA15, Q9H0Q7 | Gene names | C1orf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf49. | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.012214 (rank : 53) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
ZNFX1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.007769 (rank : 75) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.014060 (rank : 45) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.012021 (rank : 55) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.012009 (rank : 56) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.013240 (rank : 47) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.013824 (rank : 46) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.010315 (rank : 64) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
RNF8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.009151 (rank : 69) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 622 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O76064 | Gene names | RNF8, KIAA0646 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase protein RNF8 (EC 6.3.2.-) (RING finger protein 8). | |||||
|
TXD11_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.012623 (rank : 50) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6PKC3, O95887, Q6PJA6, Q8N2Q4, Q96K45, Q96K53 | Gene names | TXNDC11, EFP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11 (EF-hand-binding protein 1). | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.007869 (rank : 74) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CI039_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.012224 (rank : 52) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
DPM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.058284 (rank : 39) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60762, O15157 | Gene names | DPM1 | |||
|
Domain Architecture |

|
|||||
| Description | Dolichol-phosphate mannosyltransferase (EC 2.4.1.83) (Dolichol- phosphate mannose synthase) (Dolichyl-phosphate beta-D- mannosyltransferase) (Mannose-P-dolichol synthase) (MPD synthase) (DPM synthase). | |||||
|
DPM1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.090189 (rank : 37) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O70152, Q9D829 | Gene names | Dpm1 | |||
|
Domain Architecture |

|
|||||
| Description | Dolichol-phosphate mannosyltransferase (EC 2.4.1.83) (Dolichol- phosphate mannose synthase) (Dolichyl-phosphate beta-D- mannosyltransferase) (Mannose-P-dolichol synthase) (MPD synthase) (DPM synthase). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.011713 (rank : 57) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.011442 (rank : 61) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.009038 (rank : 70) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CLUL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.012213 (rank : 54) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15846 | Gene names | CLUL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-like protein 1 precursor (Retinal-specific clusterin-like protein). | |||||
|
CI093_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.012468 (rank : 51) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.001168 (rank : 82) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.000935 (rank : 83) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.000703 (rank : 85) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.000752 (rank : 84) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.011617 (rank : 59) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.011013 (rank : 63) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
STX4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.007999 (rank : 73) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70452 | Gene names | Stx4, Stx4a | |||
|
Domain Architecture |
|
|||||
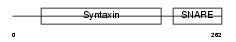
| Description | Syntaxin-4. | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.008663 (rank : 71) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
DHX57_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.001899 (rank : 81) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.010010 (rank : 65) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
KIF4A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.007007 (rank : 78) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.009788 (rank : 67) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.009868 (rank : 66) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.011619 (rank : 58) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.011393 (rank : 62) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.004101 (rank : 80) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.009465 (rank : 68) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
HMCN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | -0.004366 (rank : 86) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 983 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RW7, Q5TYR7, Q96DN3, Q96DN8, Q96SC3 | Gene names | HMCN1, FIBL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemicentin-1 precursor (Fibulin-6) (FIBL-6). | |||||
|
KIF4A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.007355 (rank : 77) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.008059 (rank : 72) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
NDEL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.013118 (rank : 48) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9GZM8, Q86T80, Q8TAR7, Q9UH50 | Gene names | NDEL1, EOPA, MITAP1, NUDEL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE-like 1 (Protein Nudel) (Mitosin- associated protein 1). | |||||
|
NDEL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.013069 (rank : 49) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ERR1, Q9D0Q4, Q9EPT6 | Gene names | Ndel1, Nudel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE-like 1 (Protein Nudel) (Protein mNudE-like) (mNudE-L). | |||||
|
SAS10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.005323 (rank : 79) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JI13, Q3UL95, Q8R5C6, Q9JJ12 | Gene names | Crlz1, Crl1, Sas10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Something about silencing protein 10 (Disrupter of silencing SAS10) (Charged amino acid-rich leucine zipper 1) (Crl-1). | |||||
|
B4GN4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.060299 (rank : 38) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
GALT8_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9NY28 | Gene names | GALNT8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable polypeptide N-acetylgalactosaminyltransferase 8 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 8) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 8) (Polypeptide GalNAc transferase 8) (GalNAc-T8) (pp-GaNTase 8). | |||||
|
GLTL4_HUMAN
|
||||||
| NC score | 0.993910 (rank : 2) | θ value | 5.94893e-161 (rank : 2) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6P9A2, O95903, Q8NDY9 | Gene names | GALNTL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 4) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 4) (Polypeptide GalNAc transferase-like protein 4) (GalNAc-T-like protein 4) (pp-GaNTase-like protein 4). | |||||
|
GALT9_HUMAN
|
||||||
| NC score | 0.993803 (rank : 3) | θ value | 4.73554e-142 (rank : 4) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HCQ5, Q6NT54, Q8NFR1 | Gene names | GALNT9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 9 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 9) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 9) (Polypeptide GalNAc transferase 9) (GalNAc-T9) (pp-GaNTase 9). | |||||
|
GLTL4_MOUSE
|
||||||
| NC score | 0.993593 (rank : 4) | θ value | 6.1562e-158 (rank : 3) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K1B9 | Gene names | Galntl4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 4) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 4) (Polypeptide GalNAc transferase-like protein 4) (GalNAc-T-like protein 4) (pp-GaNTase-like protein 4). | |||||
|
GLTL3_HUMAN
|
||||||
| NC score | 0.992529 (rank : 5) | θ value | 7.56041e-140 (rank : 5) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6IS24, Q8NFV9, Q9NTA8 | Gene names | WBSCR17, GALNTL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 3) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 3) (Polypeptide GalNAc transferase-like protein 3) (GalNAc-T-like protein 3) (pp-GaNTase-like protein 3) (Williams Beuren syndrome chromosome region 17 protein). | |||||
|
GLTL3_MOUSE
|
||||||
| NC score | 0.992451 (rank : 6) | θ value | 2.19974e-139 (rank : 6) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TT15, Q8BKN7, Q8BUY1, Q8BX73, Q8BZC8, Q8K483 | Gene names | Wbscr17, Galntl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 3) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 3) (Polypeptide GalNAc transferase-like protein 3) (GalNAc-T-like protein 3) (pp-GaNTase-like protein 3) (Williams-Beuren syndrome chromosome region 17 protein homolog). | |||||
|
GLTL5_MOUSE
|
||||||
| NC score | 0.979839 (rank : 7) | θ value | 3.34864e-71 (rank : 17) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D4M9, Q810S4, Q9CW06 | Gene names | Galntl5, Galnt15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 5 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 15) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 15) (Polypeptide GalNAc transferase 15) (GalNAc-T15) (pp-GaNTase 15). | |||||
|
GLT11_HUMAN
|
||||||
| NC score | 0.977464 (rank : 8) | θ value | 3.0217e-80 (rank : 8) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8NCW6, Q6PCD1, Q9H6C2, Q9H6Z5, Q9UDR8 | Gene names | GALNT11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 11 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 11) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 11) (Polypeptide GalNAc transferase 11) (GalNAc-T11) (pp-GaNTase 11). | |||||
|
GLTL5_HUMAN
|
||||||
| NC score | 0.977455 (rank : 9) | θ value | 1.2325e-65 (rank : 30) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z4T8, Q75KN2, Q75MD3, Q8NCV4, Q8WW05, Q9UDR9 | Gene names | GALNTL5, GALNT15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 5 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 15) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 15) (Polypeptide GalNAc transferase 15) (GalNAc-T15) (pp-GaNTase 15). | |||||
|
GLT11_MOUSE
|
||||||
| NC score | 0.977406 (rank : 10) | θ value | 4.36332e-79 (rank : 11) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q921L8, Q3TT83, Q8BU26, Q8K032 | Gene names | Galnt11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 11 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 11) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 11) (Polypeptide GalNAc transferase 11) (GalNAc-T11) (pp-GaNTase 11). | |||||
|
GALT1_MOUSE
|
||||||
| NC score | 0.975826 (rank : 11) | θ value | 5.69867e-79 (rank : 12) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O08912, Q5BKS3, Q7TND1 | Gene names | Galnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 1 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 1) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 1) (Polypeptide GalNAc transferase 1) (GalNAc-T1) (pp-GaNTase 1) [Contains: Polypeptide N-acetylgalactosaminyltransferase 1 soluble form]. | |||||
|
GALT1_HUMAN
|
||||||
| NC score | 0.975527 (rank : 12) | θ value | 2.55803e-79 (rank : 10) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q10472, Q86TJ7, Q9UM86 | Gene names | GALNT1 | |||
|
Domain Architecture |

|
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 1 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 1) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 1) (Polypeptide GalNAc transferase 1) (GalNAc-T1) (pp-GaNTase 1) [Contains: Polypeptide N-acetylgalactosaminyltransferase 1 soluble form]. | |||||
|
GLT13_HUMAN
|
||||||
| NC score | 0.975513 (rank : 13) | θ value | 2.31364e-80 (rank : 7) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IUC8, Q6ZWG1, Q96PX0, Q9UIE5 | Gene names | GALNT13, KIAA1918 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 13 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 13) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 13) (Polypeptide GalNAc transferase 13) (GalNAc-T13) (pp-GaNTase 13). | |||||
|
GLT13_MOUSE
|
||||||
| NC score | 0.975509 (rank : 14) | θ value | 3.94648e-80 (rank : 9) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CF93, Q8BLE4, Q8BYT3 | Gene names | Galnt13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 13 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 13) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 13) (Polypeptide GalNAc transferase 13) (GalNAc-T13) (pp-GaNTase 13). | |||||
|
GALT4_MOUSE
|
||||||
| NC score | 0.974317 (rank : 15) | θ value | 7.44272e-79 (rank : 13) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O08832, Q3U681 | Gene names | Galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 4) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 4) (Polypeptide GalNAc transferase 4) (GalNAc-T4) (pp-GaNTase 4). | |||||
|
GALT4_HUMAN
|
||||||
| NC score | 0.974237 (rank : 16) | θ value | 2.02684e-76 (rank : 14) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N4A0, O00208 | Gene names | GALNT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 4 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 4) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 4) (Polypeptide GalNAc transferase 4) (GalNAc-T4) (pp-GaNTase 4). | |||||
|
GALT2_MOUSE
|
||||||
| NC score | 0.973926 (rank : 17) | θ value | 5.71191e-71 (rank : 19) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6PB93, Q7TSI5, Q8BL27, Q922K5, Q99ME1 | Gene names | Galnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 2 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 2) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 2) (Polypeptide GalNAc transferase 2) (GalNAc-T2) (pp-GaNTase 2) [Contains: Polypeptide N-acetylgalactosaminyltransferase 2 soluble form]. | |||||
|
GALT2_HUMAN
|
||||||
| NC score | 0.973911 (rank : 18) | θ value | 5.71191e-71 (rank : 18) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q10471, Q9NPY4 | Gene names | GALNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 2 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 2) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 2) (Polypeptide GalNAc transferase 2) (GalNAc-T2) (pp-GaNTase 2) [Contains: Polypeptide N-acetylgalactosaminyltransferase 2 soluble form]. | |||||
|
GLT12_MOUSE
|
||||||
| NC score | 0.973723 (rank : 19) | θ value | 7.98882e-65 (rank : 32) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BGT9 | Gene names | Galnt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 12 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 12) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 12) (Polypeptide GalNAc transferase 12) (GalNAc-T12) (pp-GaNTase 12). | |||||
|
GLT12_HUMAN
|
||||||
| NC score | 0.973648 (rank : 20) | θ value | 4.23606e-66 (rank : 27) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IXK2, Q5TCF7, Q8NG54, Q96CT9, Q9H771 | Gene names | GALNT12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 12 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 12) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 12) (Polypeptide GalNAc transferase 12) (GalNAc-T12) (pp-GaNTase 12). | |||||
|
GLT10_MOUSE
|
||||||
| NC score | 0.973548 (rank : 21) | θ value | 9.43692e-66 (rank : 28) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6P9S7, Q6KAQ2, Q8BZU8, Q91YJ6 | Gene names | Galnt10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 10 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 10) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 10) (Polypeptide GalNAc transferase 10) (GalNAc-T10) (pp-GaNTase 10). | |||||
|
GLT10_HUMAN
|
||||||
| NC score | 0.973441 (rank : 22) | θ value | 1.6097e-65 (rank : 31) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q86SR1, Q6IN56, Q86VP8, Q8IXJ2, Q8TEJ2, Q96IV2, Q9H8E1, Q9Y4M4 | Gene names | GALNT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 10 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 10) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 10) (Polypeptide GalNAc transferase 10) (GalNAc-T10) (pp-GaNTase 10). | |||||
|
GLTL1_HUMAN
|
||||||
| NC score | 0.972312 (rank : 23) | θ value | 8.24801e-70 (rank : 20) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N428, Q9ULT9 | Gene names | GALNTL1, KIAA1130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 1 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 1) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 1) (Polypeptide GalNAc transferase-like protein 1) (GalNAc-T-like protein 1) (pp-GaNTase-like protein 1). | |||||
|
GLTL1_MOUSE
|
||||||
| NC score | 0.972284 (rank : 24) | θ value | 4.09345e-69 (rank : 23) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JJ61 | Gene names | Galntl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 1 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 1) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 1) (Polypeptide GalNAc transferase-like protein 1) (GalNAc-T-like protein 1) (pp-GaNTase-like protein 1). | |||||
|
GLTL2_HUMAN
|
||||||
| NC score | 0.972149 (rank : 25) | θ value | 9.43692e-66 (rank : 29) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N3T1, Q86T60, Q96C46, Q96DJ5 | Gene names | GALNTL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase-like protein 2 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 2) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 2) (Polypeptide GalNAc transferase-like protein 2) (GalNAc-T-like protein 2) (pp-GaNTase-like protein 2). | |||||
|
GLT14_HUMAN
|
||||||
| NC score | 0.971398 (rank : 26) | θ value | 1.11475e-66 (rank : 26) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96FL9, Q8IVI4, Q9BRH1, Q9H827, Q9H9J8 | Gene names | GALNT14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 14 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 14) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 14) (Polypeptide GalNAc transferase 14) (GalNAc-T14) (pp-GaNTase 14). | |||||
|
GALT6_MOUSE
|
||||||
| NC score | 0.970849 (rank : 27) | θ value | 4.67263e-73 (rank : 15) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8C7U7, Q3TWF0, Q8CED2, Q9QZ16 | Gene names | Galnt6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
GLT14_MOUSE
|
||||||
| NC score | 0.970793 (rank : 28) | θ value | 1.04338e-64 (rank : 33) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BVG5, Q60GT2, Q8BTI6, Q9DCJ2 | Gene names | Galnt14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 14 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 14) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 14) (Polypeptide GalNAc transferase 14) (GalNAc-T14) (pp-GaNTase 14). | |||||
|
GALT6_HUMAN
|
||||||
| NC score | 0.970743 (rank : 29) | θ value | 1.04096e-72 (rank : 16) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NCL4, Q8IYH4, Q9H6G2, Q9UIV5 | Gene names | GALNT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
GALT5_MOUSE
|
||||||
| NC score | 0.970583 (rank : 30) | θ value | 5.34618e-69 (rank : 24) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8C102 | Gene names | Galnt5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
GALT7_HUMAN
|
||||||
| NC score | 0.970319 (rank : 31) | θ value | 2.56992e-63 (rank : 34) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q86SF2, Q7Z5W7, Q9UJ28 | Gene names | GALNT7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylgalactosaminyltransferase 7 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 7) (UDP-GalNAc:polypeptide N- acetylgalactosaminyltransferase 7) (Polypeptide GalNAc transferase 7) (GalNAc-T7) (pp-GaNTase 7). | |||||
|
GALT7_MOUSE
|
||||||
| NC score | 0.970160 (rank : 32) | θ value | 1.66577e-62 (rank : 35) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80VA0, Q8BY62, Q8BZ70, Q8BZW0, Q91VW6, Q99MD7 | Gene names | Galnt7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylgalactosaminyltransferase 7 (EC 2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 7) (UDP-GalNAc:polypeptide N- acetylgalactosaminyltransferase 7) (Polypeptide GalNAc transferase 7) (GalNAc-T7) (pp-GaNTase 7). | |||||
|
GALT3_MOUSE
|
||||||
| NC score | 0.969456 (rank : 33) | θ value | 6.98233e-69 (rank : 25) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70419, Q80V55 | Gene names | Galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 3) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 3) (Polypeptide GalNAc transferase 3) (GalNAc-T3) (pp-GaNTase 3). | |||||
|
GALT3_HUMAN
|
||||||
| NC score | 0.969192 (rank : 34) | θ value | 1.40689e-69 (rank : 21) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q14435, Q7Z476 | Gene names | GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 3 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 3) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 3) (Polypeptide GalNAc transferase 3) (GalNAc-T3) (pp-GaNTase 3). | |||||
|
GLTL2_MOUSE
|
||||||
| NC score | 0.968998 (rank : 35) | θ value | 3.974e-56 (rank : 36) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D2N8 | Gene names | Galntl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase-like protein 2 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase-like protein 2) (UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase- like protein 2) (Polypeptide GalNAc transferase-like protein 2) (GalNAc-T-like protein 2) (pp-GaNTase-like protein 2). | |||||
|
GALT5_HUMAN
|
||||||
| NC score | 0.968463 (rank : 36) | θ value | 1.40689e-69 (rank : 22) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z7M9, Q9UGK7, Q9UHL6 | Gene names | GALNT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
DPM1_MOUSE
|
||||||
| NC score | 0.090189 (rank : 37) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O70152, Q9D829 | Gene names | Dpm1 | |||
|
Domain Architecture |

|
|||||
| Description | Dolichol-phosphate mannosyltransferase (EC 2.4.1.83) (Dolichol- phosphate mannose synthase) (Dolichyl-phosphate beta-D- mannosyltransferase) (Mannose-P-dolichol synthase) (MPD synthase) (DPM synthase). | |||||
|
B4GN4_MOUSE
|
||||||
| NC score | 0.060299 (rank : 38) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q766D5, Q8CHV2 | Gene names | B4galnt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 1 (EC 2.4.1.244) (NGalNAc-T1) (Beta- 1,4-N-acetylgalactosaminyltransferase IV) (Beta4GalNAc-T4) (Beta4GalNAcT4). | |||||
|
DPM1_HUMAN
|
||||||
| NC score | 0.058284 (rank : 39) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60762, O15157 | Gene names | DPM1 | |||
|
Domain Architecture |

|
|||||
| Description | Dolichol-phosphate mannosyltransferase (EC 2.4.1.83) (Dolichol- phosphate mannose synthase) (Dolichyl-phosphate beta-D- mannosyltransferase) (Mannose-P-dolichol synthase) (MPD synthase) (DPM synthase). | |||||
|
CT096_HUMAN
|
||||||
| NC score | 0.023315 (rank : 40) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NUD7, Q8N840, Q8NAX5 | Gene names | C20orf96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf96. | |||||
|
CA049_HUMAN
|
||||||
| NC score | 0.019160 (rank : 41) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5T0J7, Q5T0J9, Q8IYU6, Q8N7Z1, Q8NA15, Q9H0Q7 | Gene names | C1orf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf49. | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.019123 (rank : 42) | θ value | 0.163984 (rank : 37) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.014831 (rank : 43) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.014593 (rank : 44) | θ value | 0.279714 (rank : 38) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.014060 (rank : 45) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.013824 (rank : 46) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.013240 (rank : 47) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
NDEL1_HUMAN
|
||||||
| NC score | 0.013118 (rank : 48) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9GZM8, Q86T80, Q8TAR7, Q9UH50 | Gene names | NDEL1, EOPA, MITAP1, NUDEL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE-like 1 (Protein Nudel) (Mitosin- associated protein 1). | |||||
|
NDEL1_MOUSE
|
||||||
| NC score | 0.013069 (rank : 49) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ERR1, Q9D0Q4, Q9EPT6 | Gene names | Ndel1, Nudel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE-like 1 (Protein Nudel) (Protein mNudE-like) (mNudE-L). | |||||
|
TXD11_HUMAN
|
||||||
| NC score | 0.012623 (rank : 50) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6PKC3, O95887, Q6PJA6, Q8N2Q4, Q96K45, Q96K53 | Gene names | TXNDC11, EFP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11 (EF-hand-binding protein 1). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.012468 (rank : 51) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.012224 (rank : 52) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.012214 (rank : 53) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
CLUL1_HUMAN
|
||||||
| NC score | 0.012213 (rank : 54) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15846 | Gene names | CLUL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-like protein 1 precursor (Retinal-specific clusterin-like protein). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.012021 (rank : 55) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.012009 (rank : 56) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.011713 (rank : 57) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.011619 (rank : 58) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.011617 (rank : 59) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.011468 (rank : 60) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.011442 (rank : 61) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.011393 (rank : 62) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.011013 (rank : 63) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.010315 (rank : 64) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.010010 (rank : 65) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.009868 (rank : 66) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.009788 (rank : 67) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.009465 (rank : 68) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
RNF8_HUMAN
|
||||||
| NC score | 0.009151 (rank : 69) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 622 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O76064 | Gene names | RNF8, KIAA0646 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase protein RNF8 (EC 6.3.2.-) (RING finger protein 8). | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.009038 (rank : 70) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.008663 (rank : 71) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.008059 (rank : 72) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
STX4_MOUSE
|
||||||
| NC score | 0.007999 (rank : 73) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70452 | Gene names | Stx4, Stx4a | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-4. | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.007869 (rank : 74) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
ZNFX1_HUMAN
|
||||||
| NC score | 0.007769 (rank : 75) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
RAI14_HUMAN
|
||||||
| NC score | 0.007448 (rank : 76) | θ value | 0.365318 (rank : 39) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
KIF4A_MOUSE
|
||||||
| NC score | 0.007355 (rank : 77) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
KIF4A_HUMAN
|
||||||
| NC score | 0.007007 (rank : 78) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
SAS10_MOUSE
|
||||||
| NC score | 0.005323 (rank : 79) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JI13, Q3UL95, Q8R5C6, Q9JJ12 | Gene names | Crlz1, Crl1, Sas10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Something about silencing protein 10 (Disrupter of silencing SAS10) (Charged amino acid-rich leucine zipper 1) (Crl-1). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.004101 (rank : 80) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
DHX57_MOUSE
|
||||||
| NC score | 0.001899 (rank : 81) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.001168 (rank : 82) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.000935 (rank : 83) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | 0.000752 (rank : 84) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.000703 (rank : 85) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
HMCN1_HUMAN
|
||||||
| NC score | -0.004366 (rank : 86) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 983 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RW7, Q5TYR7, Q96DN3, Q96DN8, Q96SC3 | Gene names | HMCN1, FIBL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemicentin-1 precursor (Fibulin-6) (FIBL-6). | |||||