Please be patient as the page loads
|
LFNG_MOUSE
|
||||||
| SwissProt Accessions | O09010, Q3U659, Q8K3F1, Q9DC10 | Gene names | Lfng | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase lunatic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LFNG_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O09010, Q3U659, Q8K3F1, Q9DC10 | Gene names | Lfng | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase lunatic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
LFNG_HUMAN
|
||||||
| θ value | 7.7336e-177 (rank : 2) | NC score | 0.997803 (rank : 2) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NES3, O00589, Q96C39, Q9UJW5 | Gene names | LFNG | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase lunatic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
RFNG_HUMAN
|
||||||
| θ value | 4.33309e-103 (rank : 3) | NC score | 0.989298 (rank : 3) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y644, O00588 | Gene names | RFNG | |||
|
Domain Architecture |
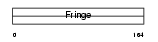
|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase radical fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
MFNG_HUMAN
|
||||||
| θ value | 2.62879e-100 (rank : 4) | NC score | 0.980841 (rank : 6) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00587, O43730 | Gene names | MFNG | |||
|
Domain Architecture |
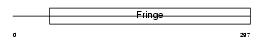
|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase manic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
RFNG_MOUSE
|
||||||
| θ value | 4.48402e-100 (rank : 5) | NC score | 0.986206 (rank : 4) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09009 | Gene names | Rfng | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase radical fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
MFNG_MOUSE
|
||||||
| θ value | 1.10445e-98 (rank : 6) | NC score | 0.981760 (rank : 5) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09008 | Gene names | Mfng | |||
|
Domain Architecture |
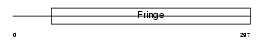
|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase manic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
B3GLT_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 7) | NC score | 0.480409 (rank : 7) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6Y288, Q5W0H2, Q6NUI3 | Gene names | B3GALTL, B3GTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
B3GLT_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 8) | NC score | 0.473171 (rank : 8) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHT6, Q0PCR7, Q149S5 | Gene names | B3galtl, Gm1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
CHSS1_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 9) | NC score | 0.261832 (rank : 9) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS1_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 10) | NC score | 0.248258 (rank : 10) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86X52, Q6UX38, Q7LFU5, Q9Y2J5 | Gene names | CHSY1, CHSY, CSS1, KIAA0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase 1) (Chondroitin sulfate synthase 1) (N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS3_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 11) | NC score | 0.237310 (rank : 11) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
CHGUT_HUMAN
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.179639 (rank : 12) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2E5, Q6P2I4, Q6UXD2 | Gene names | CSGLCAT, KIAA1402 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate glucuronyltransferase (EC 2.4.1.226) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (Chondroitin glucuronyltransferase II) (CSGlcA-T). | |||||
|
CRSP7_HUMAN
|
||||||
| θ value | 0.813845 (rank : 13) | NC score | 0.034988 (rank : 19) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
COBA2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.009543 (rank : 22) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13942, Q07751, Q13271, Q13272, Q13273, Q99866, Q9UIP9 | Gene names | COL11A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
SMRD3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.011068 (rank : 21) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P9Z1, Q921D3, Q9CX89, Q9DB99 | Gene names | Smarcd3, Baf60c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 3 (60 kDa BRG-1/Brm-associated factor subunit C) (BRG1-associated factor 60C) (mBAF60c). | |||||
|
B3GL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.015465 (rank : 20) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75752, Q3Y531, Q6IAI5, Q8NFM8, Q9HA06 | Gene names | B3GALNT1, B3GALT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.79) (Beta-3-GalNAc-T1) (Beta-1,3-galactosyltransferase 3) (Beta-1,3- GalTase 3) (Beta3Gal-T3) (b3Gal-T3) (Galactosylgalactosylglucosylceramide beta-D-acetyl- galactosaminyltransferase) (UDP-N- acetylgalactosamine:globotriaosylceramide beta-1,3-N- acetylgalactosaminyltransferase) (Globoside synthase) (Beta-3-Gx-T3). | |||||
|
COBA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.007739 (rank : 24) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.008525 (rank : 23) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.003246 (rank : 25) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
IKBE_MOUSE
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.002603 (rank : 26) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54910, Q3U686, Q9CZZ9, Q9D7U3 | Gene names | Nfkbie, Ikbe | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKB-E). | |||||
|
CGAT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 21) | NC score | 0.060208 (rank : 15) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TDX6, Q6P9G6, Q8IUF9, Q9NSQ7, Q9NUM9 | Gene names | GALNACT1, CHGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 22) | NC score | 0.060075 (rank : 16) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BJQ9, Q3UZX6, Q8BWV9, Q8BZU7, Q8C195, Q8R0M6 | Gene names | Galnact1, Chgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.059339 (rank : 18) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N6G5, Q6MZJ5, Q6MZP6, Q8TCH4, Q9P1I6 | Gene names | GALNACT2, CHGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CGAT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 24) | NC score | 0.059454 (rank : 17) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C1F4, Q6PAP6, Q8R5A2, Q9D2M1 | Gene names | Galnact2, Chgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CHSS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 25) | NC score | 0.069934 (rank : 14) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IZ52, Q6UXD6, Q7L4G1, Q9H0F8, Q9H618 | Gene names | CSS2, CHPF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase) (Chondroitin-polymerizing factor). | |||||
|
CHSS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 26) | NC score | 0.071976 (rank : 13) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6IQX7 | Gene names | Css2, D1Bwg1363e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase). | |||||
|
LFNG_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O09010, Q3U659, Q8K3F1, Q9DC10 | Gene names | Lfng | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase lunatic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
LFNG_HUMAN
|
||||||
| NC score | 0.997803 (rank : 2) | θ value | 7.7336e-177 (rank : 2) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NES3, O00589, Q96C39, Q9UJW5 | Gene names | LFNG | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase lunatic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
RFNG_HUMAN
|
||||||
| NC score | 0.989298 (rank : 3) | θ value | 4.33309e-103 (rank : 3) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y644, O00588 | Gene names | RFNG | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase radical fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
RFNG_MOUSE
|
||||||
| NC score | 0.986206 (rank : 4) | θ value | 4.48402e-100 (rank : 5) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09009 | Gene names | Rfng | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase radical fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
MFNG_MOUSE
|
||||||
| NC score | 0.981760 (rank : 5) | θ value | 1.10445e-98 (rank : 6) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09008 | Gene names | Mfng | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase manic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
MFNG_HUMAN
|
||||||
| NC score | 0.980841 (rank : 6) | θ value | 2.62879e-100 (rank : 4) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00587, O43730 | Gene names | MFNG | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-N-acetylglucosaminyltransferase manic fringe (EC 2.4.1.222) (O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase). | |||||
|
B3GLT_HUMAN
|
||||||
| NC score | 0.480409 (rank : 7) | θ value | 6.21693e-09 (rank : 7) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6Y288, Q5W0H2, Q6NUI3 | Gene names | B3GALTL, B3GTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
B3GLT_MOUSE
|
||||||
| NC score | 0.473171 (rank : 8) | θ value | 6.87365e-08 (rank : 8) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHT6, Q0PCR7, Q149S5 | Gene names | B3galtl, Gm1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,3-glucosyltransferase (EC 2.4.1.-) (Beta3Glc-T) (Beta-3- glycosyltransferase-like). | |||||
|
CHSS1_MOUSE
|
||||||
| NC score | 0.261832 (rank : 9) | θ value | 2.21117e-06 (rank : 9) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZQ11 | Gene names | Chsy1, Kiaa0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase) (Chondroitin sulfate synthase 1) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS1_HUMAN
|
||||||
| NC score | 0.248258 (rank : 10) | θ value | 1.09739e-05 (rank : 10) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86X52, Q6UX38, Q7LFU5, Q9Y2J5 | Gene names | CHSY1, CHSY, CSS1, KIAA0990 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 1 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase 1) (Chondroitin sulfate synthase 1) (N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase 1) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II). | |||||
|
CHSS3_HUMAN
|
||||||
| NC score | 0.237310 (rank : 11) | θ value | 0.000121331 (rank : 11) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
CHGUT_HUMAN
|
||||||
| NC score | 0.179639 (rank : 12) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P2E5, Q6P2I4, Q6UXD2 | Gene names | CSGLCAT, KIAA1402 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate glucuronyltransferase (EC 2.4.1.226) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (Chondroitin glucuronyltransferase II) (CSGlcA-T). | |||||
|
CHSS2_MOUSE
|
||||||
| NC score | 0.071976 (rank : 13) | θ value | θ > 10 (rank : 26) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6IQX7 | Gene names | Css2, D1Bwg1363e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase). | |||||
|
CHSS2_HUMAN
|
||||||
| NC score | 0.069934 (rank : 14) | θ value | θ > 10 (rank : 25) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IZ52, Q6UXD6, Q7L4G1, Q9H0F8, Q9H618 | Gene names | CSS2, CHPF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 2 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (N-acetylgalactosaminyl- proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase) (Chondroitin-polymerizing factor). | |||||
|
CGAT1_HUMAN
|
||||||
| NC score | 0.060208 (rank : 15) | θ value | θ > 10 (rank : 21) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TDX6, Q6P9G6, Q8IUF9, Q9NSQ7, Q9NUM9 | Gene names | GALNACT1, CHGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT1_MOUSE
|
||||||
| NC score | 0.060075 (rank : 16) | θ value | θ > 10 (rank : 22) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BJQ9, Q3UZX6, Q8BWV9, Q8BZU7, Q8C195, Q8R0M6 | Gene names | Galnact1, Chgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.174) (beta4GalNAcT-1). | |||||
|
CGAT2_MOUSE
|
||||||
| NC score | 0.059454 (rank : 17) | θ value | θ > 10 (rank : 24) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C1F4, Q6PAP6, Q8R5A2, Q9D2M1 | Gene names | Galnact2, Chgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CGAT2_HUMAN
|
||||||
| NC score | 0.059339 (rank : 18) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N6G5, Q6MZJ5, Q6MZP6, Q8TCH4, Q9P1I6 | Gene names | GALNACT2, CHGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin beta-1,4-N-acetylgalactosaminyltransferase 2 (EC 2.4.1.174) (GalNAcT-2) (beta4GalNAcT-2). | |||||
|
CRSP7_HUMAN
|
||||||
| NC score | 0.034988 (rank : 19) | θ value | 0.813845 (rank : 13) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
B3GL1_HUMAN
|
||||||
| NC score | 0.015465 (rank : 20) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75752, Q3Y531, Q6IAI5, Q8NFM8, Q9HA06 | Gene names | B3GALNT1, B3GALT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.79) (Beta-3-GalNAc-T1) (Beta-1,3-galactosyltransferase 3) (Beta-1,3- GalTase 3) (Beta3Gal-T3) (b3Gal-T3) (Galactosylgalactosylglucosylceramide beta-D-acetyl- galactosaminyltransferase) (UDP-N- acetylgalactosamine:globotriaosylceramide beta-1,3-N- acetylgalactosaminyltransferase) (Globoside synthase) (Beta-3-Gx-T3). | |||||
|
SMRD3_MOUSE
|
||||||
| NC score | 0.011068 (rank : 21) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P9Z1, Q921D3, Q9CX89, Q9DB99 | Gene names | Smarcd3, Baf60c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 3 (60 kDa BRG-1/Brm-associated factor subunit C) (BRG1-associated factor 60C) (mBAF60c). | |||||
|
COBA2_HUMAN
|
||||||
| NC score | 0.009543 (rank : 22) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13942, Q07751, Q13271, Q13272, Q13273, Q99866, Q9UIP9 | Gene names | COL11A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.008525 (rank : 23) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
COBA2_MOUSE
|
||||||
| NC score | 0.007739 (rank : 24) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.003246 (rank : 25) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
IKBE_MOUSE
|
||||||
| NC score | 0.002603 (rank : 26) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54910, Q3U686, Q9CZZ9, Q9D7U3 | Gene names | Nfkbie, Ikbe | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKB-E). | |||||