Please be patient as the page loads
|
MYCB_MOUSE
|
||||||
| SwissProt Accessions | Q6P8Z1, Q9D6E3 | Gene names | Mycb, Bmyc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein B-Myc. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MYCB_MOUSE
|
||||||
| θ value | 1.82472e-93 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6P8Z1, Q9D6E3 | Gene names | Mycb, Bmyc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein B-Myc. | |||||
|
MYC_MOUSE
|
||||||
| θ value | 1.85029e-45 (rank : 2) | NC score | 0.830056 (rank : 2) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYC_HUMAN
|
||||||
| θ value | 3.05695e-40 (rank : 3) | NC score | 0.828112 (rank : 3) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYCN_MOUSE
|
||||||
| θ value | 1.86753e-13 (rank : 4) | NC score | 0.694378 (rank : 4) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCN_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 5) | NC score | 0.689254 (rank : 6) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCS_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 6) | NC score | 0.690598 (rank : 5) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
MYCL1_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 7) | NC score | 0.607657 (rank : 7) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL1_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 8) | NC score | 0.569292 (rank : 8) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL2_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 9) | NC score | 0.563294 (rank : 9) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
KLKB1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.015053 (rank : 52) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P26262 | Gene names | Klkb1, Klk3, Pk | |||
|
Domain Architecture |

|
|||||
| Description | Plasma kallikrein precursor (EC 3.4.21.34) (Plasma prekallikrein) (Kininogenin) (Fletcher factor) [Contains: Plasma kallikrein heavy chain; Plasma kallikrein light chain]. | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 2.36792 (rank : 11) | NC score | 0.014025 (rank : 53) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
TIF1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 12) | NC score | 0.011910 (rank : 54) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
DIAP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 13) | NC score | 0.007094 (rank : 55) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
R3HD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 14) | NC score | 0.019076 (rank : 51) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15032, Q8IW32 | Gene names | R3HDM1, KIAA0029, R3HDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R3H domain-containing protein 1. | |||||
|
ARNT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 15) | NC score | 0.051148 (rank : 50) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ASCL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 16) | NC score | 0.078144 (rank : 37) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P50553, Q9BQ30 | Gene names | ASCL1, ASH1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (HASH1). | |||||
|
ASCL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 17) | NC score | 0.070236 (rank : 41) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02067 | Gene names | Ascl1, Ash1, Mash-1, Mash1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (Mash-1). | |||||
|
ASCL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 18) | NC score | 0.085011 (rank : 32) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35885, Q9WUJ7 | Gene names | Ascl2, Mash2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash-2). | |||||
|
BHLH8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 19) | NC score | 0.093328 (rank : 31) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7RTS1 | Gene names | BHLHB8, MIST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
BHLH8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 20) | NC score | 0.104310 (rank : 30) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
FIGLA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 21) | NC score | 0.132698 (rank : 17) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6QHK4 | Gene names | FIGLA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
FIGLA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 22) | NC score | 0.147666 (rank : 13) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O55208 | Gene names | Figla | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
HAND1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.053912 (rank : 47) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64279, Q61099 | Gene names | Hand1, Ehand, Hxt, Thing1 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND) (Helix-loop-helix transcription factor expressed in extraembryonic mesoderm and trophoblast) (Thing-1) (Th1). | |||||
|
HAND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 24) | NC score | 0.060670 (rank : 43) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P61296, O95300, O95301, P97833 | Gene names | HAND2, DHAND | |||
|
Domain Architecture |
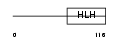
|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND). | |||||
|
HAND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 25) | NC score | 0.060670 (rank : 44) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61039, Q61100 | Gene names | Hand2, Dhand, Hed, Thing2 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND) (Helix-loop-helix transcription factor expressed in embryo and deciduum) (Thing-2). | |||||
|
MAD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 26) | NC score | 0.145098 (rank : 15) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MAD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 27) | NC score | 0.140713 (rank : 16) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
MAD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.131006 (rank : 18) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MAD4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.112696 (rank : 26) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MAX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.303469 (rank : 10) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MAX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.294758 (rank : 11) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MITF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.083275 (rank : 34) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.082144 (rank : 36) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MLX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.117011 (rank : 24) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.116107 (rank : 25) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MNT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.125212 (rank : 21) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.128126 (rank : 19) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MXI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.145856 (rank : 14) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MXI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.152237 (rank : 12) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
PTF1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.076429 (rank : 38) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7RTS3, Q9HC25 | Gene names | PTF1A, PTF1P48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
PTF1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.064200 (rank : 42) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QX98, Q9QYF5, Q9QYF6 | Gene names | Ptf1a, Ptf1p48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
SRBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.082815 (rank : 35) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.074733 (rank : 40) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
SRBP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.106965 (rank : 29) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
TFAP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.123587 (rank : 22) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.083601 (rank : 33) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
TFEB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.056308 (rank : 46) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.056647 (rank : 45) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TWST2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.052710 (rank : 49) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WVJ9 | Gene names | TWIST2, DERMO1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.052791 (rank : 48) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D030 | Gene names | Twist2, Dermo1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
USF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.122770 (rank : 23) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.127856 (rank : 20) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.110954 (rank : 27) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.110536 (rank : 28) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
WBS14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.075703 (rank : 39) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
MYCB_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.82472e-93 (rank : 1) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6P8Z1, Q9D6E3 | Gene names | Mycb, Bmyc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein B-Myc. | |||||
|
MYC_MOUSE
|
||||||
| NC score | 0.830056 (rank : 2) | θ value | 1.85029e-45 (rank : 2) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYC_HUMAN
|
||||||
| NC score | 0.828112 (rank : 3) | θ value | 3.05695e-40 (rank : 3) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYCN_MOUSE
|
||||||
| NC score | 0.694378 (rank : 4) | θ value | 1.86753e-13 (rank : 4) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCS_MOUSE
|
||||||
| NC score | 0.690598 (rank : 5) | θ value | 4.30538e-10 (rank : 6) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
MYCN_HUMAN
|
||||||
| NC score | 0.689254 (rank : 6) | θ value | 2.43908e-13 (rank : 5) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCL1_HUMAN
|
||||||
| NC score | 0.607657 (rank : 7) | θ value | 1.29631e-06 (rank : 7) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL1_MOUSE
|
||||||
| NC score | 0.569292 (rank : 8) | θ value | 2.21117e-06 (rank : 8) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL2_HUMAN
|
||||||
| NC score | 0.563294 (rank : 9) | θ value | 0.000602161 (rank : 9) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
MAX_HUMAN
|
||||||
| NC score | 0.303469 (rank : 10) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MAX_MOUSE
|
||||||
| NC score | 0.294758 (rank : 11) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MXI1_MOUSE
|
||||||
| NC score | 0.152237 (rank : 12) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
FIGLA_MOUSE
|
||||||
| NC score | 0.147666 (rank : 13) | θ value | θ > 10 (rank : 22) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O55208 | Gene names | Figla | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
MXI1_HUMAN
|
||||||
| NC score | 0.145856 (rank : 14) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MAD3_HUMAN
|
||||||
| NC score | 0.145098 (rank : 15) | θ value | θ > 10 (rank : 26) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MAD3_MOUSE
|
||||||
| NC score | 0.140713 (rank : 16) | θ value | θ > 10 (rank : 27) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
FIGLA_HUMAN
|
||||||
| NC score | 0.132698 (rank : 17) | θ value | θ > 10 (rank : 21) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6QHK4 | Gene names | FIGLA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
MAD4_HUMAN
|
||||||
| NC score | 0.131006 (rank : 18) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.128126 (rank : 19) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.127856 (rank : 20) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.125212 (rank : 21) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
TFAP4_HUMAN
|
||||||
| NC score | 0.123587 (rank : 22) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.122770 (rank : 23) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.117011 (rank : 24) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.116107 (rank : 25) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MAD4_MOUSE
|
||||||
| NC score | 0.112696 (rank : 26) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.110954 (rank : 27) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| NC score | 0.110536 (rank : 28) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
SRBP2_HUMAN
|
||||||
| NC score | 0.106965 (rank : 29) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
BHLH8_MOUSE
|
||||||
| NC score | 0.104310 (rank : 30) | θ value | θ > 10 (rank : 20) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
BHLH8_HUMAN
|
||||||
| NC score | 0.093328 (rank : 31) | θ value | θ > 10 (rank : 19) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7RTS1 | Gene names | BHLHB8, MIST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
ASCL2_MOUSE
|
||||||
| NC score | 0.085011 (rank : 32) | θ value | θ > 10 (rank : 18) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35885, Q9WUJ7 | Gene names | Ascl2, Mash2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash-2). | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.083601 (rank : 33) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.083275 (rank : 34) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
SRBP1_HUMAN
|
||||||
| NC score | 0.082815 (rank : 35) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.082144 (rank : 36) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
ASCL1_HUMAN
|
||||||
| NC score | 0.078144 (rank : 37) | θ value | θ > 10 (rank : 16) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P50553, Q9BQ30 | Gene names | ASCL1, ASH1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (HASH1). | |||||
|
PTF1A_HUMAN
|
||||||
| NC score | 0.076429 (rank : 38) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7RTS3, Q9HC25 | Gene names | PTF1A, PTF1P48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
WBS14_HUMAN
|
||||||
| NC score | 0.075703 (rank : 39) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.074733 (rank : 40) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
ASCL1_MOUSE
|
||||||
| NC score | 0.070236 (rank : 41) | θ value | θ > 10 (rank : 17) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02067 | Gene names | Ascl1, Ash1, Mash-1, Mash1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (Mash-1). | |||||
|
PTF1A_MOUSE
|
||||||
| NC score | 0.064200 (rank : 42) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QX98, Q9QYF5, Q9QYF6 | Gene names | Ptf1a, Ptf1p48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
HAND2_HUMAN
|
||||||
| NC score | 0.060670 (rank : 43) | θ value | θ > 10 (rank : 24) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P61296, O95300, O95301, P97833 | Gene names | HAND2, DHAND | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND). | |||||
|
HAND2_MOUSE
|
||||||
| NC score | 0.060670 (rank : 44) | θ value | θ > 10 (rank : 25) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61039, Q61100 | Gene names | Hand2, Dhand, Hed, Thing2 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND) (Helix-loop-helix transcription factor expressed in embryo and deciduum) (Thing-2). | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.056647 (rank : 45) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.056308 (rank : 46) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
HAND1_MOUSE
|
||||||
| NC score | 0.053912 (rank : 47) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64279, Q61099 | Gene names | Hand1, Ehand, Hxt, Thing1 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND) (Helix-loop-helix transcription factor expressed in extraembryonic mesoderm and trophoblast) (Thing-1) (Th1). | |||||
|
TWST2_MOUSE
|
||||||
| NC score | 0.052791 (rank : 48) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D030 | Gene names | Twist2, Dermo1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST2_HUMAN
|
||||||
| NC score | 0.052710 (rank : 49) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WVJ9 | Gene names | TWIST2, DERMO1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.051148 (rank : 50) | θ value | θ > 10 (rank : 15) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
R3HD1_HUMAN
|
||||||
| NC score | 0.019076 (rank : 51) | θ value | 6.88961 (rank : 14) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15032, Q8IW32 | Gene names | R3HDM1, KIAA0029, R3HDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R3H domain-containing protein 1. | |||||
|
KLKB1_MOUSE
|
||||||
| NC score | 0.015053 (rank : 52) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P26262 | Gene names | Klkb1, Klk3, Pk | |||
|
Domain Architecture |

|
|||||
| Description | Plasma kallikrein precursor (EC 3.4.21.34) (Plasma prekallikrein) (Kininogenin) (Fletcher factor) [Contains: Plasma kallikrein heavy chain; Plasma kallikrein light chain]. | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.014025 (rank : 53) | θ value | 2.36792 (rank : 11) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
TIF1A_MOUSE
|
||||||
| NC score | 0.011910 (rank : 54) | θ value | 4.03905 (rank : 12) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
DIAP1_MOUSE
|
||||||
| NC score | 0.007094 (rank : 55) | θ value | 6.88961 (rank : 13) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||