Please be patient as the page loads
|
MYCL2_HUMAN
|
||||||
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MYCL2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
MYCL1_HUMAN
|
||||||
| θ value | 1.13238e-127 (rank : 2) | NC score | 0.944511 (rank : 2) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL1_MOUSE
|
||||||
| θ value | 1.2549e-118 (rank : 3) | NC score | 0.938542 (rank : 3) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCN_MOUSE
|
||||||
| θ value | 4.43474e-23 (rank : 4) | NC score | 0.745339 (rank : 4) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCN_HUMAN
|
||||||
| θ value | 7.56453e-23 (rank : 5) | NC score | 0.743515 (rank : 5) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCS_MOUSE
|
||||||
| θ value | 2.27762e-19 (rank : 6) | NC score | 0.716184 (rank : 6) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
MYC_MOUSE
|
||||||
| θ value | 1.47631e-18 (rank : 7) | NC score | 0.712192 (rank : 8) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYC_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 8) | NC score | 0.715749 (rank : 7) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MAX_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 9) | NC score | 0.473922 (rank : 11) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MAX_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 10) | NC score | 0.477860 (rank : 10) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MAD3_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 11) | NC score | 0.322557 (rank : 14) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MAD3_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 12) | NC score | 0.311234 (rank : 15) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
MYCB_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 13) | NC score | 0.563294 (rank : 9) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P8Z1, Q9D6E3 | Gene names | Mycb, Bmyc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein B-Myc. | |||||
|
MXI1_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 14) | NC score | 0.343227 (rank : 12) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
WBS14_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 15) | NC score | 0.184373 (rank : 30) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
MXI1_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 16) | NC score | 0.331716 (rank : 13) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MAD4_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 17) | NC score | 0.308661 (rank : 16) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MAD4_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 18) | NC score | 0.276291 (rank : 17) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MAD1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 19) | NC score | 0.219959 (rank : 20) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
NDF2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 20) | NC score | 0.088897 (rank : 50) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15784, Q8TBI7, Q9UQC6 | Gene names | NEUROD2, NDRF | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
NDF2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 21) | NC score | 0.088414 (rank : 52) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62414, Q61952, Q925V5 | Gene names | Neurod2, Ndrf | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 22) | NC score | 0.214758 (rank : 21) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
MITF_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 23) | NC score | 0.195983 (rank : 27) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 24) | NC score | 0.193340 (rank : 28) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
KIF5C_HUMAN
|
||||||
| θ value | 0.125558 (rank : 25) | NC score | 0.019221 (rank : 81) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
USF2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 26) | NC score | 0.225708 (rank : 19) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 27) | NC score | 0.226476 (rank : 18) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
KIF5C_MOUSE
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.018562 (rank : 82) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
FIGLA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.202124 (rank : 26) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6QHK4 | Gene names | FIGLA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
WBS14_MOUSE
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.117644 (rank : 42) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.021560 (rank : 78) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SIGIR_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.029775 (rank : 75) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IA17, Q3KQY2, Q6UXI3, Q9H733 | Gene names | SIGIRR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Single Ig IL-1-related receptor (Single Ig IL-1R-related molecule) (Single immunoglobulin domain-containing IL1R-related protein) (Toll/interleukin-1 receptor 8) (TIR8). | |||||
|
NDF1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.059692 (rank : 65) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
T22D2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.031045 (rank : 74) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
TJAP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.026434 (rank : 76) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DCD5, Q8CFL7, Q8R5I2 | Gene names | Tjap1, Pilt, Tjp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.002373 (rank : 94) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
FIGLA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.191538 (rank : 29) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O55208 | Gene names | Figla | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
TMAP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.057720 (rank : 66) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CZH7, Q8C6F8 | Gene names | Tmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
MELPH_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.022230 (rank : 77) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BV36, Q9HA71 | Gene names | MLPH, SLAC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Synaptotagmin-like protein 2a) (Slp homolog lacking C2 domains a). | |||||
|
NDF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.056101 (rank : 67) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
RBM6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.019999 (rank : 80) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
TFEB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.155368 (rank : 35) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.156604 (rank : 34) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
MINT_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.020878 (rank : 79) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.007965 (rank : 91) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
PDXL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.017674 (rank : 83) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZ53, Q6UVY4, Q8WUV6 | Gene names | PODXL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
SCN3A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.005454 (rank : 92) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.011645 (rank : 85) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.091801 (rank : 47) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.089262 (rank : 49) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
BHLH8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.135029 (rank : 39) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7RTS1 | Gene names | BHLHB8, MIST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
BHLH8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.142936 (rank : 37) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
FOXGB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.004143 (rank : 93) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55315 | Gene names | FOXG1B, FKHL1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein G1B (Forkhead-related protein FKHL1) (Transcription factor BF-1) (Brain factor 1) (BF1) (HFK1). | |||||
|
GLU2B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.012874 (rank : 84) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08795, Q921X2 | Gene names | Prkcsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.010188 (rank : 87) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.009788 (rank : 88) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
U2AFL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.010321 (rank : 86) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
GGA3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.008336 (rank : 89) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
HAND1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.077417 (rank : 57) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O96004 | Gene names | HAND1, EHAND | |||
|
Domain Architecture |
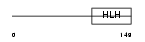
|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND). | |||||
|
HAND1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.081872 (rank : 54) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q64279, Q61099 | Gene names | Hand1, Ehand, Hxt, Thing1 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND) (Helix-loop-helix transcription factor expressed in extraembryonic mesoderm and trophoblast) (Thing-1) (Th1). | |||||
|
HAND2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.098406 (rank : 44) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P61296, O95300, O95301, P97833 | Gene names | HAND2, DHAND | |||
|
Domain Architecture |
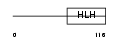
|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND). | |||||
|
HAND2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.098406 (rank : 45) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61039, Q61100 | Gene names | Hand2, Dhand, Hed, Thing2 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND) (Helix-loop-helix transcription factor expressed in embryo and deciduum) (Thing-2). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.008229 (rank : 90) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.076119 (rank : 61) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.076638 (rank : 58) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ASCL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.100564 (rank : 43) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P50553, Q9BQ30 | Gene names | ASCL1, ASH1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (HASH1). | |||||
|
ASCL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.088775 (rank : 51) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02067 | Gene names | Ascl1, Ash1, Mash-1, Mash1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (Mash-1). | |||||
|
ASCL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.063083 (rank : 64) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99929, Q9UM68 | Gene names | ASCL2 | |||
|
Domain Architecture |
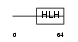
|
|||||
| Description | Achaete-scute homolog 2 (Mash2) (HASH2). | |||||
|
ASCL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.092801 (rank : 46) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O35885, Q9WUJ7 | Gene names | Ascl2, Mash2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash-2). | |||||
|
ATOH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.052450 (rank : 72) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92858 | Gene names | ATOH1, ATH1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein hATH-1). | |||||
|
ATOH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.052211 (rank : 73) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P48985 | Gene names | Atoh1, Ath1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein mATH-1) (MATH1). | |||||
|
BHLH3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.053577 (rank : 70) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
MAD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.164398 (rank : 33) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MLX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.165798 (rank : 32) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.166920 (rank : 31) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MNT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.205997 (rank : 25) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.213227 (rank : 22) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
NGN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.054975 (rank : 68) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H2A3, Q8N416 | Gene names | NEUROG2, NGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogenin 2. | |||||
|
NGN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.053465 (rank : 71) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70447, P70237 | Gene names | Neurog2, Ath4a, Atoh4, Ngn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 2 (Atonal protein homolog 4) (Helix-loop-helix protein mATH-4A) (MATH4A). | |||||
|
NGN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.073218 (rank : 62) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y4Z2, Q9BY24 | Gene names | NEUROG3, NGN3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3. | |||||
|
NGN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.054883 (rank : 69) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70661 | Gene names | Neurog3, Ath4b, Atoh5, Ngn3 | |||
|
Domain Architecture |
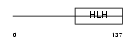
|
|||||
| Description | Neurogenin 3 (Atonal protein homolog 5) (Helix-loop-helix protein mATH-4B) (MATH4B). | |||||
|
PTF1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.089601 (rank : 48) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7RTS3, Q9HC25 | Gene names | PTF1A, PTF1P48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
PTF1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.087364 (rank : 53) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QX98, Q9QYF5, Q9QYF6 | Gene names | Ptf1a, Ptf1p48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
SCX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.072438 (rank : 63) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q64124 | Gene names | Scx | |||
|
Domain Architecture |
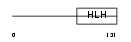
|
|||||
| Description | Basic helix-loop-helix transcription factor scleraxis. | |||||
|
SRBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.127698 (rank : 40) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.119167 (rank : 41) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
SRBP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.137927 (rank : 38) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
TFAP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.149720 (rank : 36) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
TWST1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.076583 (rank : 59) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15672, Q92487, Q99804 | Gene names | TWIST1, TWIST | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (H-twist). | |||||
|
TWST1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.076503 (rank : 60) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26687 | Gene names | Twist1, Twist | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (M-twist). | |||||
|
TWST2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.081713 (rank : 56) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WVJ9 | Gene names | TWIST2, DERMO1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.081838 (rank : 55) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D030 | Gene names | Twist2, Dermo1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
USF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.206737 (rank : 24) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.208848 (rank : 23) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
MYCL2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
MYCL1_HUMAN
|
||||||
| NC score | 0.944511 (rank : 2) | θ value | 1.13238e-127 (rank : 2) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL1_MOUSE
|
||||||
| NC score | 0.938542 (rank : 3) | θ value | 1.2549e-118 (rank : 3) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCN_MOUSE
|
||||||
| NC score | 0.745339 (rank : 4) | θ value | 4.43474e-23 (rank : 4) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCN_HUMAN
|
||||||
| NC score | 0.743515 (rank : 5) | θ value | 7.56453e-23 (rank : 5) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCS_MOUSE
|
||||||
| NC score | 0.716184 (rank : 6) | θ value | 2.27762e-19 (rank : 6) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
MYC_HUMAN
|
||||||
| NC score | 0.715749 (rank : 7) | θ value | 1.63225e-17 (rank : 8) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYC_MOUSE
|
||||||
| NC score | 0.712192 (rank : 8) | θ value | 1.47631e-18 (rank : 7) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYCB_MOUSE
|
||||||
| NC score | 0.563294 (rank : 9) | θ value | 0.000602161 (rank : 13) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P8Z1, Q9D6E3 | Gene names | Mycb, Bmyc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein B-Myc. | |||||
|
MAX_HUMAN
|
||||||
| NC score | 0.477860 (rank : 10) | θ value | 3.19293e-05 (rank : 10) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MAX_MOUSE
|
||||||
| NC score | 0.473922 (rank : 11) | θ value | 1.87187e-05 (rank : 9) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MXI1_MOUSE
|
||||||
| NC score | 0.343227 (rank : 12) | θ value | 0.000786445 (rank : 14) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MXI1_HUMAN
|
||||||
| NC score | 0.331716 (rank : 13) | θ value | 0.00298849 (rank : 16) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MAD3_HUMAN
|
||||||
| NC score | 0.322557 (rank : 14) | θ value | 0.00020696 (rank : 11) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MAD3_MOUSE
|
||||||
| NC score | 0.311234 (rank : 15) | θ value | 0.000461057 (rank : 12) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
MAD4_HUMAN
|
||||||
| NC score | 0.308661 (rank : 16) | θ value | 0.00509761 (rank : 17) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MAD4_MOUSE
|
||||||
| NC score | 0.276291 (rank : 17) | θ value | 0.0148317 (rank : 18) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
USF2_MOUSE
|
||||||
| NC score | 0.226476 (rank : 18) | θ value | 0.125558 (rank : 27) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.225708 (rank : 19) | θ value | 0.125558 (rank : 26) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
MAD1_MOUSE
|
||||||
| NC score | 0.219959 (rank : 20) | θ value | 0.0330416 (rank : 19) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.214758 (rank : 21) | θ value | 0.0431538 (rank : 22) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.213227 (rank : 22) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.208848 (rank : 23) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.206737 (rank : 24) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.205997 (rank : 25) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
FIGLA_HUMAN
|
||||||
| NC score | 0.202124 (rank : 26) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6QHK4 | Gene names | FIGLA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.195983 (rank : 27) | θ value | 0.0736092 (rank : 23) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.193340 (rank : 28) | θ value | 0.0961366 (rank : 24) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
FIGLA_MOUSE
|
||||||
| NC score | 0.191538 (rank : 29) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O55208 | Gene names | Figla | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
WBS14_HUMAN
|
||||||
| NC score | 0.184373 (rank : 30) | θ value | 0.00228821 (rank : 15) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.166920 (rank : 31) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.165798 (rank : 32) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MAD1_HUMAN
|
||||||
| NC score | 0.164398 (rank : 33) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.156604 (rank : 34) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.155368 (rank : 35) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFAP4_HUMAN
|
||||||
| NC score | 0.149720 (rank : 36) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
BHLH8_MOUSE
|
||||||
| NC score | 0.142936 (rank : 37) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
SRBP2_HUMAN
|
||||||
| NC score | 0.137927 (rank : 38) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
BHLH8_HUMAN
|
||||||
| NC score | 0.135029 (rank : 39) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7RTS1 | Gene names | BHLHB8, MIST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
SRBP1_HUMAN
|
||||||
| NC score | 0.127698 (rank : 40) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.119167 (rank : 41) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
WBS14_MOUSE
|
||||||
| NC score | 0.117644 (rank : 42) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
ASCL1_HUMAN
|
||||||
| NC score | 0.100564 (rank : 43) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P50553, Q9BQ30 | Gene names | ASCL1, ASH1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (HASH1). | |||||
|
HAND2_HUMAN
|
||||||
| NC score | 0.098406 (rank : 44) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P61296, O95300, O95301, P97833 | Gene names | HAND2, DHAND | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND). | |||||
|
HAND2_MOUSE
|
||||||
| NC score | 0.098406 (rank : 45) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61039, Q61100 | Gene names | Hand2, Dhand, Hed, Thing2 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND) (Helix-loop-helix transcription factor expressed in embryo and deciduum) (Thing-2). | |||||
|
ASCL2_MOUSE
|
||||||
| NC score | 0.092801 (rank : 46) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O35885, Q9WUJ7 | Gene names | Ascl2, Mash2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash-2). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.091801 (rank : 47) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
PTF1A_HUMAN
|
||||||
| NC score | 0.089601 (rank : 48) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7RTS3, Q9HC25 | Gene names | PTF1A, PTF1P48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.089262 (rank : 49) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NDF2_HUMAN
|
||||||
| NC score | 0.088897 (rank : 50) | θ value | 0.0330416 (rank : 20) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15784, Q8TBI7, Q9UQC6 | Gene names | NEUROD2, NDRF | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
ASCL1_MOUSE
|
||||||
| NC score | 0.088775 (rank : 51) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02067 | Gene names | Ascl1, Ash1, Mash-1, Mash1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (Mash-1). | |||||
|
NDF2_MOUSE
|
||||||
| NC score | 0.088414 (rank : 52) | θ value | 0.0431538 (rank : 21) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62414, Q61952, Q925V5 | Gene names | Neurod2, Ndrf | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
PTF1A_MOUSE
|
||||||
| NC score | 0.087364 (rank : 53) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QX98, Q9QYF5, Q9QYF6 | Gene names | Ptf1a, Ptf1p48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
HAND1_MOUSE
|
||||||
| NC score | 0.081872 (rank : 54) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q64279, Q61099 | Gene names | Hand1, Ehand, Hxt, Thing1 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND) (Helix-loop-helix transcription factor expressed in extraembryonic mesoderm and trophoblast) (Thing-1) (Th1). | |||||
|
TWST2_MOUSE
|
||||||
| NC score | 0.081838 (rank : 55) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D030 | Gene names | Twist2, Dermo1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST2_HUMAN
|
||||||
| NC score | 0.081713 (rank : 56) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WVJ9 | Gene names | TWIST2, DERMO1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
HAND1_HUMAN
|
||||||
| NC score | 0.077417 (rank : 57) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O96004 | Gene names | HAND1, EHAND | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND). | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.076638 (rank : 58) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
TWST1_HUMAN
|
||||||
| NC score | 0.076583 (rank : 59) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15672, Q92487, Q99804 | Gene names | TWIST1, TWIST | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (H-twist). | |||||
|
TWST1_MOUSE
|
||||||
| NC score | 0.076503 (rank : 60) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26687 | Gene names | Twist1, Twist | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (M-twist). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.076119 (rank : 61) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
NGN3_HUMAN
|
||||||
| NC score | 0.073218 (rank : 62) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y4Z2, Q9BY24 | Gene names | NEUROG3, NGN3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3. | |||||
|
SCX_MOUSE
|
||||||
| NC score | 0.072438 (rank : 63) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q64124 | Gene names | Scx | |||
|
Domain Architecture |

|
|||||
| Description | Basic helix-loop-helix transcription factor scleraxis. | |||||
|
ASCL2_HUMAN
|
||||||
| NC score | 0.063083 (rank : 64) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99929, Q9UM68 | Gene names | ASCL2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash2) (HASH2). | |||||
|
NDF1_MOUSE
|
||||||
| NC score | 0.059692 (rank : 65) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
TMAP1_MOUSE
|
||||||
| NC score | 0.057720 (rank : 66) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CZH7, Q8C6F8 | Gene names | Tmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
NDF1_HUMAN
|
||||||
| NC score | 0.056101 (rank : 67) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
NGN2_HUMAN
|
||||||
| NC score | 0.054975 (rank : 68) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H2A3, Q8N416 | Gene names | NEUROG2, NGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogenin 2. | |||||
|
NGN3_MOUSE
|
||||||
| NC score | 0.054883 (rank : 69) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70661 | Gene names | Neurog3, Ath4b, Atoh5, Ngn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3 (Atonal protein homolog 5) (Helix-loop-helix protein mATH-4B) (MATH4B). | |||||
|
BHLH3_HUMAN
|
||||||
| NC score | 0.053577 (rank : 70) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
NGN2_MOUSE
|
||||||
| NC score | 0.053465 (rank : 71) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70447, P70237 | Gene names | Neurog2, Ath4a, Atoh4, Ngn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 2 (Atonal protein homolog 4) (Helix-loop-helix protein mATH-4A) (MATH4A). | |||||
|
ATOH1_HUMAN
|
||||||
| NC score | 0.052450 (rank : 72) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92858 | Gene names | ATOH1, ATH1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein hATH-1). | |||||
|
ATOH1_MOUSE
|
||||||
| NC score | 0.052211 (rank : 73) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P48985 | Gene names | Atoh1, Ath1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein mATH-1) (MATH1). | |||||
|
T22D2_HUMAN
|
||||||
| NC score | 0.031045 (rank : 74) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
SIGIR_HUMAN
|
||||||
| NC score | 0.029775 (rank : 75) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IA17, Q3KQY2, Q6UXI3, Q9H733 | Gene names | SIGIRR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Single Ig IL-1-related receptor (Single Ig IL-1R-related molecule) (Single immunoglobulin domain-containing IL1R-related protein) (Toll/interleukin-1 receptor 8) (TIR8). | |||||
|
TJAP1_MOUSE
|
||||||
| NC score | 0.026434 (rank : 76) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DCD5, Q8CFL7, Q8R5I2 | Gene names | Tjap1, Pilt, Tjp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
MELPH_HUMAN
|
||||||
| NC score | 0.022230 (rank : 77) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BV36, Q9HA71 | Gene names | MLPH, SLAC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Synaptotagmin-like protein 2a) (Slp homolog lacking C2 domains a). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.021560 (rank : 78) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.020878 (rank : 79) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RBM6_HUMAN
|
||||||
| NC score | 0.019999 (rank : 80) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
KIF5C_HUMAN
|
||||||
| NC score | 0.019221 (rank : 81) | θ value | 0.125558 (rank : 25) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
KIF5C_MOUSE
|
||||||
| NC score | 0.018562 (rank : 82) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
PDXL2_HUMAN
|
||||||
| NC score | 0.017674 (rank : 83) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZ53, Q6UVY4, Q8WUV6 | Gene names | PODXL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
GLU2B_MOUSE
|
||||||
| NC score | 0.012874 (rank : 84) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08795, Q921X2 | Gene names | Prkcsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.011645 (rank : 85) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
U2AFL_HUMAN
|
||||||
| NC score | 0.010321 (rank : 86) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.010188 (rank : 87) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.009788 (rank : 88) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
GGA3_MOUSE
|
||||||
| NC score | 0.008336 (rank : 89) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.008229 (rank : 90) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.007965 (rank : 91) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
SCN3A_HUMAN
|
||||||
| NC score | 0.005454 (rank : 92) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
FOXGB_HUMAN
|
||||||
| NC score | 0.004143 (rank : 93) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55315 | Gene names | FOXG1B, FKHL1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein G1B (Forkhead-related protein FKHL1) (Transcription factor BF-1) (Brain factor 1) (BF1) (HFK1). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.002373 (rank : 94) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||