Please be patient as the page loads
|
DEAF1_MOUSE
|
||||||
| SwissProt Accessions | Q9Z1T5 | Gene names | Deaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DEAF1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.992106 (rank : 2) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
DEAF1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9Z1T5 | Gene names | Deaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR). | |||||
|
MTG8_HUMAN
|
||||||
| θ value | 1.29331e-14 (rank : 3) | NC score | 0.528834 (rank : 4) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q06455, Q06456, Q14873, Q16239, Q16346, Q16347, Q92479, Q9BRZ0 | Gene names | RUNX1T1, AML1T1, CBFA2T1, CDR, ETO, MTG8, ZMYND2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8) (Protein ETO) (Eight twenty one protein) (Cyclin-D-related protein) (Zinc finger MYND domain- containing protein 2). | |||||
|
MTG8_MOUSE
|
||||||
| θ value | 1.29331e-14 (rank : 4) | NC score | 0.529492 (rank : 3) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61909 | Gene names | Runx1t1, Cbfa2t1, Cbfa2t1h, Mtg8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8). | |||||
|
SP100_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 5) | NC score | 0.310011 (rank : 14) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P23497, O75450, Q13343, Q96F70, Q96T24, Q96T95, Q9NP33, Q9UE32 | Gene names | SP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein) (Lysp100b). | |||||
|
MTG8R_MOUSE
|
||||||
| θ value | 1.133e-10 (rank : 6) | NC score | 0.491948 (rank : 5) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70374 | Gene names | Cbfa2t2, Cbfa2t2h, Mtgr1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1). | |||||
|
LY10_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 7) | NC score | 0.373628 (rank : 8) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
MTG8R_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 8) | NC score | 0.488723 (rank : 6) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43439, Q5TGE4, Q5TGE7, Q8IWF3, Q96B06, Q96L00, Q9H436, Q9UJP8, Q9UJP9, Q9UP24 | Gene names | CBFA2T2, EHT, MTGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1) (Myeloid translocation-related protein 1) (ETO homologous on chromosome 20) (p85). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 9) | NC score | 0.288489 (rank : 17) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
IPR1_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 10) | NC score | 0.442624 (rank : 7) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BVK9, Q3UCV7, Q80V00 | Gene names | Ipr1, Ifi75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intracellular pathogen resistance protein 1. | |||||
|
SP110_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 11) | NC score | 0.356428 (rank : 9) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
GMEB2_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 12) | NC score | 0.339768 (rank : 10) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P58929, Q6PCY0 | Gene names | Gmeb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucocorticoid modulatory element-binding protein 2 (GMEB-2). | |||||
|
GMEB2_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 13) | NC score | 0.334511 (rank : 11) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKD1, Q5TDS0, Q9H431, Q9H4X7, Q9H4X8, Q9UF78, Q9ULF1 | Gene names | GMEB2, KIAA1269 | |||
|
Domain Architecture |
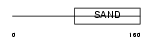
|
|||||
| Description | Glucocorticoid modulatory element-binding protein 2 (GMEB-2) (Parvovirus initiation factor p79) (PIF p79) (DNA-binding protein p79PIF). | |||||
|
GMEB1_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 14) | NC score | 0.313362 (rank : 12) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JL60 | Gene names | Gmeb1 | |||
|
Domain Architecture |
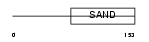
|
|||||
| Description | Glucocorticoid modulatory element-binding protein 1 (GMEB-1). | |||||
|
GMEB1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 15) | NC score | 0.311096 (rank : 13) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y692, Q9NWH1, Q9UKD0 | Gene names | GMEB1 | |||
|
Domain Architecture |
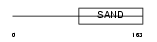
|
|||||
| Description | Glucocorticoid modulatory element-binding protein 1 (GMEB-1) (Parvovirus initiation factor p96) (PIF p96) (DNA-binding protein p96PIF). | |||||
|
ZMY11_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 16) | NC score | 0.270796 (rank : 19) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
SP100_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 17) | NC score | 0.308028 (rank : 16) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35892, O35897, O88392, O88395 | Gene names | Sp100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein). | |||||
|
ZMY11_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 18) | NC score | 0.263696 (rank : 20) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
SMYD3_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 19) | NC score | 0.242587 (rank : 23) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9CWR2, Q6P7V6, Q8BG90 | Gene names | Smyd3, Zmynd1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
SMYD3_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 20) | NC score | 0.244046 (rank : 22) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H7B4, Q86TL8, Q8N5Z6, Q96AI5 | Gene names | SMYD3, ZMYND1, ZNFN3A1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
EGLN1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 21) | NC score | 0.192119 (rank : 29) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9GZT9, Q8N3M8, Q9BZS8, Q9BZT0 | Gene names | EGLN1, C1orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (Prolyl hydroxylase domain-containing protein 2) (PHD2) (SM-20). | |||||
|
ZMY17_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 22) | NC score | 0.309794 (rank : 15) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9D5Z5 | Gene names | Zmynd17 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 17. | |||||
|
EGLN1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 23) | NC score | 0.185869 (rank : 30) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91YE3, Q8VHJ2, Q922P3 | Gene names | Egln1 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (SM-20). | |||||
|
ZMY10_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 24) | NC score | 0.276993 (rank : 18) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99ML0, Q80W73 | Gene names | Zmynd10, Blu | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 10 (BLu protein). | |||||
|
ANKY2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 25) | NC score | 0.133249 (rank : 36) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IV38, Q96BL3 | Gene names | ANKMY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
SMYD2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 26) | NC score | 0.201330 (rank : 27) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R5A0 | Gene names | Smyd2 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 2. | |||||
|
ZMY10_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 27) | NC score | 0.262868 (rank : 21) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75800, O14570, O75801, Q8N4R6, Q8NDN6 | Gene names | ZMYND10, BLU | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 10 (BLu protein). | |||||
|
ANKY1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 28) | NC score | 0.086969 (rank : 39) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P2S6, Q8IYX5, Q8NDK5, Q9NX10 | Gene names | ANKMY1, TSAL1, ZMYND13 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 1 (Testis-specific ankyrin-like protein 1) (Zinc-finger MYND domain-containing protein 13). | |||||
|
ANKY2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 29) | NC score | 0.135968 (rank : 35) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q3TPE9, Q8BK14, Q8BYW5, Q8R3N4, Q921J1 | Gene names | Ankmy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
SMYD2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 30) | NC score | 0.203399 (rank : 26) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NRG4, Q4V765, Q5VSH9 | Gene names | SMYD2 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 2 (HSKM-B). | |||||
|
PDCD2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 31) | NC score | 0.173737 (rank : 31) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q16342, Q9UH12 | Gene names | PDCD2, RP8, ZMYND7 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8) (Zinc finger MYND domain-containing protein 7). | |||||
|
AIRE_MOUSE
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.120574 (rank : 37) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.020073 (rank : 54) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.014366 (rank : 57) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
PDCD2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 35) | NC score | 0.160105 (rank : 32) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P46718, Q8BR06 | Gene names | Pdcd2, Rp8 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8). | |||||
|
TM108_MOUSE
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.054473 (rank : 43) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
PDC6I_HUMAN
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.045117 (rank : 46) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
ZMY19_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.215104 (rank : 24) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96E35 | Gene names | ZMYND19, MIZIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 19 (Melanin-concentrating hormone receptor 1-interacting zinc-finger protein) (MCH-R1- interacting zinc-finger protein). | |||||
|
ZMY19_MOUSE
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.215104 (rank : 25) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9CQG3, Q8K1T3 | Gene names | Zmynd19, Mizip | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 19 (Melanin-concentrating hormone receptor 1-interacting zinc-finger protein) (MCH-R1- interacting zinc-finger protein). | |||||
|
ZN447_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.006358 (rank : 68) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
GPS2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.041349 (rank : 47) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13227 | Gene names | GPS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein pathway suppressor 2 (Protein GPS2). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.035164 (rank : 48) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
ANR44_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.010895 (rank : 65) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N8A2, Q6P480, Q86VL5, Q8IZ72, Q9UFA4 | Gene names | ANKRD44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 44. | |||||
|
MAZ_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.002001 (rank : 73) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P56270, Q15703, Q99443 | Gene names | MAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myc-associated zinc finger protein (MAZI) (Purine-binding transcription factor) (Pur-1) (ZF87) (ZIF87). | |||||
|
SMYD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.143865 (rank : 33) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NB12 | Gene names | SMYD1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 1. | |||||
|
SMYD1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.143745 (rank : 34) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97443, P97442, P97444 | Gene names | Smyd1, Bop | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 1 (Zinc-finger protein BOP) (m- BOP) (CD8b-opposite). | |||||
|
ODO2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.022464 (rank : 53) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P36957, Q9BQ32 | Gene names | DLST, DLTS | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
PDC6I_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.033592 (rank : 50) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
WASF4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.017714 (rank : 55) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
WASL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.026967 (rank : 51) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
ZCHC8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.033711 (rank : 49) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
AIRE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.097877 (rank : 38) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
ELF4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.010683 (rank : 66) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
KLF10_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.001880 (rank : 74) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O89091 | Gene names | Klf10, Gdnfif, Tieg, Tieg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 10 (Transforming growth factor-beta-inducible early growth response protein 1) (TGFB-inducible early growth response protein 1) (TIEG-1) (GDNF-inducible factor) (Transcription factor GIF) (mGIF). | |||||
|
GLI2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.001334 (rank : 75) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
RFX1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.012341 (rank : 62) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
SEM4B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.005499 (rank : 69) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPR2, Q6UXE3, Q8WVP9, Q96FK5, Q9C0B8, Q9H691, Q9NPM8, Q9NPN0 | Gene names | SEMA4B, KIAA1745 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4B precursor. | |||||
|
GCX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.023506 (rank : 52) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96NM4, Q5TE33, Q5TE34, Q5TE35, Q96IC9, Q9BQN5 | Gene names | GCX1, C20orf100 | |||
|
Domain Architecture |

|
|||||
| Description | Granulosa cell HMG box protein 1 (GCX-1). | |||||
|
RIN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.012757 (rank : 60) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TB24, Q8NF30, Q8TEE8, Q8WYP4, Q9H6A5, Q9HAG1 | Gene names | RIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
SNX2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.009212 (rank : 67) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CWK8, Q3UKW3, Q9CQV0 | Gene names | Snx2 | |||
|
Domain Architecture |
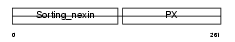
|
|||||
| Description | Sorting nexin-2. | |||||
|
SOX7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.012385 (rank : 61) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
AP180_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.011360 (rank : 63) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61548, Q61547 | Gene names | Snap91 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein) (Phosphoprotein F1- 20). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.002629 (rank : 71) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CMTA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.010935 (rank : 64) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94983, Q7Z6M8, Q8N3V0, Q8NDG4, Q96G17 | Gene names | CAMTA2, KIAA0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
FLRT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.002652 (rank : 70) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43155 | Gene names | FLRT2, KIAA0405 | |||
|
Domain Architecture |
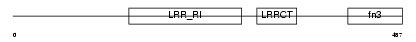
|
|||||
| Description | Leucine-rich repeat transmembrane protein FLRT2 precursor (Fibronectin-like domain-containing leucine-rich transmembrane protein 2). | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.002248 (rank : 72) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
PRDM5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | -0.000314 (rank : 76) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NQX1 | Gene names | PRDM5, PFM2 | |||
|
Domain Architecture |
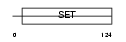
|
|||||
| Description | PR domain zinc finger protein 5 (PR domain-containing protein 5). | |||||
|
SMR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.017288 (rank : 56) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
SOX21_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.014343 (rank : 58) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y651, P35715, Q15504 | Gene names | SOX21, SOX25, SOXA | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-21 (SOX-A). | |||||
|
SOX21_MOUSE
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.014341 (rank : 59) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q811W0, Q8VH35 | Gene names | Sox21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-21. | |||||
|
EGLN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.050049 (rank : 45) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H6Z9 | Gene names | EGLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 3 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 3) (HIF-prolyl hydroxylase 3) (HIF-PH3) (HPH-1) (Prolyl hydroxylase domain-containing protein 3) (PHD3). | |||||
|
S100R_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.195893 (rank : 28) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99388 | Gene names | D1Lub1, Sp100-rs | |||
|
Domain Architecture |
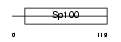
|
|||||
| Description | Putative Sp100-related protein. | |||||
|
SMYD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.061104 (rank : 41) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
SMYD4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.053524 (rank : 44) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.055593 (rank : 42) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
ZMY12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.066326 (rank : 40) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H0C1, Q5VUS6, Q8TC87, Q96M51 | Gene names | ZMYND12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 12. | |||||
|
DEAF1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9Z1T5 | Gene names | Deaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR). | |||||
|
DEAF1_HUMAN
|
||||||
| NC score | 0.992106 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
MTG8_MOUSE
|
||||||
| NC score | 0.529492 (rank : 3) | θ value | 1.29331e-14 (rank : 4) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61909 | Gene names | Runx1t1, Cbfa2t1, Cbfa2t1h, Mtg8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8). | |||||
|
MTG8_HUMAN
|
||||||
| NC score | 0.528834 (rank : 4) | θ value | 1.29331e-14 (rank : 3) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q06455, Q06456, Q14873, Q16239, Q16346, Q16347, Q92479, Q9BRZ0 | Gene names | RUNX1T1, AML1T1, CBFA2T1, CDR, ETO, MTG8, ZMYND2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8) (Protein ETO) (Eight twenty one protein) (Cyclin-D-related protein) (Zinc finger MYND domain- containing protein 2). | |||||
|
MTG8R_MOUSE
|
||||||
| NC score | 0.491948 (rank : 5) | θ value | 1.133e-10 (rank : 6) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70374 | Gene names | Cbfa2t2, Cbfa2t2h, Mtgr1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1). | |||||
|
MTG8R_HUMAN
|
||||||
| NC score | 0.488723 (rank : 6) | θ value | 2.52405e-10 (rank : 8) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43439, Q5TGE4, Q5TGE7, Q8IWF3, Q96B06, Q96L00, Q9H436, Q9UJP8, Q9UJP9, Q9UP24 | Gene names | CBFA2T2, EHT, MTGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1) (Myeloid translocation-related protein 1) (ETO homologous on chromosome 20) (p85). | |||||
|
IPR1_MOUSE
|
||||||
| NC score | 0.442624 (rank : 7) | θ value | 1.06045e-08 (rank : 10) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BVK9, Q3UCV7, Q80V00 | Gene names | Ipr1, Ifi75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intracellular pathogen resistance protein 1. | |||||
|
LY10_HUMAN
|
||||||
| NC score | 0.373628 (rank : 8) | θ value | 2.52405e-10 (rank : 7) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
SP110_HUMAN
|
||||||
| NC score | 0.356428 (rank : 9) | θ value | 7.59969e-07 (rank : 11) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
GMEB2_MOUSE
|
||||||
| NC score | 0.339768 (rank : 10) | θ value | 2.21117e-06 (rank : 12) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P58929, Q6PCY0 | Gene names | Gmeb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucocorticoid modulatory element-binding protein 2 (GMEB-2). | |||||
|
GMEB2_HUMAN
|
||||||
| NC score | 0.334511 (rank : 11) | θ value | 6.43352e-06 (rank : 13) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKD1, Q5TDS0, Q9H431, Q9H4X7, Q9H4X8, Q9UF78, Q9ULF1 | Gene names | GMEB2, KIAA1269 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid modulatory element-binding protein 2 (GMEB-2) (Parvovirus initiation factor p79) (PIF p79) (DNA-binding protein p79PIF). | |||||
|
GMEB1_MOUSE
|
||||||
| NC score | 0.313362 (rank : 12) | θ value | 9.29e-05 (rank : 14) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JL60 | Gene names | Gmeb1 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid modulatory element-binding protein 1 (GMEB-1). | |||||
|
GMEB1_HUMAN
|
||||||
| NC score | 0.311096 (rank : 13) | θ value | 0.000121331 (rank : 15) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y692, Q9NWH1, Q9UKD0 | Gene names | GMEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid modulatory element-binding protein 1 (GMEB-1) (Parvovirus initiation factor p96) (PIF p96) (DNA-binding protein p96PIF). | |||||
|
SP100_HUMAN
|
||||||
| NC score | 0.310011 (rank : 14) | θ value | 6.64225e-11 (rank : 5) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P23497, O75450, Q13343, Q96F70, Q96T24, Q96T95, Q9NP33, Q9UE32 | Gene names | SP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein) (Lysp100b). | |||||
|
ZMY17_MOUSE
|
||||||
| NC score | 0.309794 (rank : 15) | θ value | 0.00228821 (rank : 22) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9D5Z5 | Gene names | Zmynd17 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 17. | |||||
|
SP100_MOUSE
|
||||||
| NC score | 0.308028 (rank : 16) | θ value | 0.000461057 (rank : 17) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35892, O35897, O88392, O88395 | Gene names | Sp100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.288489 (rank : 17) | θ value | 2.79066e-09 (rank : 9) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
ZMY10_MOUSE
|
||||||
| NC score | 0.276993 (rank : 18) | θ value | 0.0113563 (rank : 24) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99ML0, Q80W73 | Gene names | Zmynd10, Blu | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 10 (BLu protein). | |||||
|
ZMY11_HUMAN
|
||||||
| NC score | 0.270796 (rank : 19) | θ value | 0.000121331 (rank : 16) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ZMY11_MOUSE
|
||||||
| NC score | 0.263696 (rank : 20) | θ value | 0.000602161 (rank : 18) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
ZMY10_HUMAN
|
||||||
| NC score | 0.262868 (rank : 21) | θ value | 0.0431538 (rank : 27) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75800, O14570, O75801, Q8N4R6, Q8NDN6 | Gene names | ZMYND10, BLU | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 10 (BLu protein). | |||||
|
SMYD3_HUMAN
|
||||||
| NC score | 0.244046 (rank : 22) | θ value | 0.00134147 (rank : 20) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H7B4, Q86TL8, Q8N5Z6, Q96AI5 | Gene names | SMYD3, ZMYND1, ZNFN3A1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
SMYD3_MOUSE
|
||||||
| NC score | 0.242587 (rank : 23) | θ value | 0.000786445 (rank : 19) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9CWR2, Q6P7V6, Q8BG90 | Gene names | Smyd3, Zmynd1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 3 (EC 2.1.1.43) (Zinc finger MYND domain-containing protein 1). | |||||
|
ZMY19_HUMAN
|
||||||
| NC score | 0.215104 (rank : 24) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96E35 | Gene names | ZMYND19, MIZIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 19 (Melanin-concentrating hormone receptor 1-interacting zinc-finger protein) (MCH-R1- interacting zinc-finger protein). | |||||
|
ZMY19_MOUSE
|
||||||
| NC score | 0.215104 (rank : 25) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9CQG3, Q8K1T3 | Gene names | Zmynd19, Mizip | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 19 (Melanin-concentrating hormone receptor 1-interacting zinc-finger protein) (MCH-R1- interacting zinc-finger protein). | |||||
|
SMYD2_HUMAN
|
||||||
| NC score | 0.203399 (rank : 26) | θ value | 0.0961366 (rank : 30) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NRG4, Q4V765, Q5VSH9 | Gene names | SMYD2 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 2 (HSKM-B). | |||||
|
SMYD2_MOUSE
|
||||||
| NC score | 0.201330 (rank : 27) | θ value | 0.0330416 (rank : 26) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R5A0 | Gene names | Smyd2 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 2. | |||||
|
S100R_MOUSE
|
||||||
| NC score | 0.195893 (rank : 28) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99388 | Gene names | D1Lub1, Sp100-rs | |||
|
Domain Architecture |

|
|||||
| Description | Putative Sp100-related protein. | |||||
|
EGLN1_HUMAN
|
||||||
| NC score | 0.192119 (rank : 29) | θ value | 0.00228821 (rank : 21) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9GZT9, Q8N3M8, Q9BZS8, Q9BZT0 | Gene names | EGLN1, C1orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (Prolyl hydroxylase domain-containing protein 2) (PHD2) (SM-20). | |||||
|
EGLN1_MOUSE
|
||||||
| NC score | 0.185869 (rank : 30) | θ value | 0.00509761 (rank : 23) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91YE3, Q8VHJ2, Q922P3 | Gene names | Egln1 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (SM-20). | |||||
|
PDCD2_HUMAN
|
||||||
| NC score | 0.173737 (rank : 31) | θ value | 0.125558 (rank : 31) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q16342, Q9UH12 | Gene names | PDCD2, RP8, ZMYND7 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8) (Zinc finger MYND domain-containing protein 7). | |||||
|
PDCD2_MOUSE
|
||||||
| NC score | 0.160105 (rank : 32) | θ value | 0.279714 (rank : 35) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P46718, Q8BR06 | Gene names | Pdcd2, Rp8 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 2 (Zinc finger protein Rp-8). | |||||
|
SMYD1_HUMAN
|
||||||
| NC score | 0.143865 (rank : 33) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NB12 | Gene names | SMYD1 | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 1. | |||||
|
SMYD1_MOUSE
|
||||||
| NC score | 0.143745 (rank : 34) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97443, P97442, P97444 | Gene names | Smyd1, Bop | |||
|
Domain Architecture |

|
|||||
| Description | SET and MYND domain-containing protein 1 (Zinc-finger protein BOP) (m- BOP) (CD8b-opposite). | |||||
|
ANKY2_MOUSE
|
||||||
| NC score | 0.135968 (rank : 35) | θ value | 0.0736092 (rank : 29) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q3TPE9, Q8BK14, Q8BYW5, Q8R3N4, Q921J1 | Gene names | Ankmy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
ANKY2_HUMAN
|
||||||
| NC score | 0.133249 (rank : 36) | θ value | 0.0330416 (rank : 25) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IV38, Q96BL3 | Gene names | ANKMY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
AIRE_MOUSE
|
||||||
| NC score | 0.120574 (rank : 37) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
AIRE_HUMAN
|
||||||
| NC score | 0.097877 (rank : 38) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
ANKY1_HUMAN
|
||||||
| NC score | 0.086969 (rank : 39) | θ value | 0.0563607 (rank : 28) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P2S6, Q8IYX5, Q8NDK5, Q9NX10 | Gene names | ANKMY1, TSAL1, ZMYND13 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 1 (Testis-specific ankyrin-like protein 1) (Zinc-finger MYND domain-containing protein 13). | |||||
|
ZMY12_HUMAN
|
||||||
| NC score | 0.066326 (rank : 40) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H0C1, Q5VUS6, Q8TC87, Q96M51 | Gene names | ZMYND12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 12. | |||||
|
SMYD4_HUMAN
|
||||||
| NC score | 0.061104 (rank : 41) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IYR2, Q8N1P2, Q8NAT0, Q96LV4, Q96PV2 | Gene names | SMYD4, KIAA1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.055593 (rank : 42) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TM108_MOUSE
|
||||||
| NC score | 0.054473 (rank : 43) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
SMYD4_MOUSE
|
||||||
| NC score | 0.053524 (rank : 44) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
EGLN3_HUMAN
|
||||||
| NC score | 0.050049 (rank : 45) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H6Z9 | Gene names | EGLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 3 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 3) (HIF-prolyl hydroxylase 3) (HIF-PH3) (HPH-1) (Prolyl hydroxylase domain-containing protein 3) (PHD3). | |||||
|
PDC6I_HUMAN
|
||||||
| NC score | 0.045117 (rank : 46) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
GPS2_HUMAN
|
||||||
| NC score | 0.041349 (rank : 47) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13227 | Gene names | GPS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein pathway suppressor 2 (Protein GPS2). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.035164 (rank : 48) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
ZCHC8_MOUSE
|
||||||
| NC score | 0.033711 (rank : 49) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
PDC6I_MOUSE
|
||||||
| NC score | 0.033592 (rank : 50) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.026967 (rank : 51) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
GCX1_HUMAN
|
||||||
| NC score | 0.023506 (rank : 52) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96NM4, Q5TE33, Q5TE34, Q5TE35, Q96IC9, Q9BQN5 | Gene names | GCX1, C20orf100 | |||
|
Domain Architecture |

|
|||||
| Description | Granulosa cell HMG box protein 1 (GCX-1). | |||||
|
ODO2_HUMAN
|
||||||
| NC score | 0.022464 (rank : 53) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P36957, Q9BQ32 | Gene names | DLST, DLTS | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.020073 (rank : 54) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
WASF4_HUMAN
|
||||||
| NC score | 0.017714 (rank : 55) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
SMR1_MOUSE
|
||||||
| NC score | 0.017288 (rank : 56) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.014366 (rank : 57) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
SOX21_HUMAN
|
||||||
| NC score | 0.014343 (rank : 58) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y651, P35715, Q15504 | Gene names | SOX21, SOX25, SOXA | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-21 (SOX-A). | |||||
|
SOX21_MOUSE
|
||||||
| NC score | 0.014341 (rank : 59) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q811W0, Q8VH35 | Gene names | Sox21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor SOX-21. | |||||
|
RIN3_HUMAN
|
||||||
| NC score | 0.012757 (rank : 60) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TB24, Q8NF30, Q8TEE8, Q8WYP4, Q9H6A5, Q9HAG1 | Gene names | RIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
SOX7_HUMAN
|
||||||
| NC score | 0.012385 (rank : 61) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
RFX1_MOUSE
|
||||||
| NC score | 0.012341 (rank : 62) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
AP180_MOUSE
|
||||||
| NC score | 0.011360 (rank : 63) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61548, Q61547 | Gene names | Snap91 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein) (Phosphoprotein F1- 20). | |||||
|
CMTA2_HUMAN
|
||||||
| NC score | 0.010935 (rank : 64) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94983, Q7Z6M8, Q8N3V0, Q8NDG4, Q96G17 | Gene names | CAMTA2, KIAA0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
ANR44_HUMAN
|
||||||
| NC score | 0.010895 (rank : 65) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N8A2, Q6P480, Q86VL5, Q8IZ72, Q9UFA4 | Gene names | ANKRD44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 44. | |||||
|
ELF4_MOUSE
|
||||||
| NC score | 0.010683 (rank : 66) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
SNX2_MOUSE
|
||||||
| NC score | 0.009212 (rank : 67) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CWK8, Q3UKW3, Q9CQV0 | Gene names | Snx2 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-2. | |||||
|
ZN447_HUMAN
|
||||||
| NC score | 0.006358 (rank : 68) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
SEM4B_HUMAN
|
||||||
| NC score | 0.005499 (rank : 69) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPR2, Q6UXE3, Q8WVP9, Q96FK5, Q9C0B8, Q9H691, Q9NPM8, Q9NPN0 | Gene names | SEMA4B, KIAA1745 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4B precursor. | |||||
|
FLRT2_HUMAN
|
||||||
| NC score | 0.002652 (rank : 70) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43155 | Gene names | FLRT2, KIAA0405 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat transmembrane protein FLRT2 precursor (Fibronectin-like domain-containing leucine-rich transmembrane protein 2). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.002629 (rank : 71) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
LAP4_HUMAN
|
||||||
| NC score | 0.002248 (rank : 72) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
MAZ_HUMAN
|
||||||
| NC score | 0.002001 (rank : 73) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P56270, Q15703, Q99443 | Gene names | MAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myc-associated zinc finger protein (MAZI) (Purine-binding transcription factor) (Pur-1) (ZF87) (ZIF87). | |||||
|
KLF10_MOUSE
|
||||||
| NC score | 0.001880 (rank : 74) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O89091 | Gene names | Klf10, Gdnfif, Tieg, Tieg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 10 (Transforming growth factor-beta-inducible early growth response protein 1) (TGFB-inducible early growth response protein 1) (TIEG-1) (GDNF-inducible factor) (Transcription factor GIF) (mGIF). | |||||
|
GLI2_HUMAN
|
||||||
| NC score | 0.001334 (rank : 75) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
PRDM5_HUMAN
|
||||||
| NC score | -0.000314 (rank : 76) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NQX1 | Gene names | PRDM5, PFM2 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 5 (PR domain-containing protein 5). | |||||