Please be patient as the page loads
|
MBN3_HUMAN
|
||||||
| SwissProt Accessions | Q9NUK0, Q5JXN8, Q5JXN9, Q5JXP4, Q6UDQ1, Q8TAD9, Q8TAF4, Q9H0Z7, Q9UF37 | Gene names | MBNL3, CHCR, MBLX39, MBXL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein HCHCR). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MBN3_HUMAN
|
||||||
| θ value | 7.22167e-183 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NUK0, Q5JXN8, Q5JXN9, Q5JXP4, Q6UDQ1, Q8TAD9, Q8TAF4, Q9H0Z7, Q9UF37 | Gene names | MBNL3, CHCR, MBLX39, MBXL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein HCHCR). | |||||
|
MBN3_MOUSE
|
||||||
| θ value | 3.25673e-167 (rank : 2) | NC score | 0.989341 (rank : 2) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8R003 | Gene names | Mbnl3, Chcr, Mbxl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein MCHCR). | |||||
|
MBNL_MOUSE
|
||||||
| θ value | 1.02895e-112 (rank : 3) | NC score | 0.976171 (rank : 3) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JKP5 | Gene names | Mbnl1, Exp, Mbnl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
MBNL_HUMAN
|
||||||
| θ value | 6.0463e-105 (rank : 4) | NC score | 0.975617 (rank : 4) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NR56, O43311, O43797 | Gene names | MBNL1, EXP, KIAA0428, MBNL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
ZC3H3_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 5) | NC score | 0.326488 (rank : 6) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ZC3H3_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 6) | NC score | 0.327497 (rank : 5) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
MKRN1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 7) | NC score | 0.157751 (rank : 9) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
MKRN1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 8) | NC score | 0.140773 (rank : 12) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
ZC3H6_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 9) | NC score | 0.149599 (rank : 11) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.150930 (rank : 10) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
MKRN3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.120387 (rank : 18) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
CS007_HUMAN
|
||||||
| θ value | 0.21417 (rank : 12) | NC score | 0.138158 (rank : 13) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
CS007_MOUSE
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.136264 (rank : 14) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
ANX11_HUMAN
|
||||||
| θ value | 0.279714 (rank : 14) | NC score | 0.025930 (rank : 39) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50995 | Gene names | ANXA11, ANX11 | |||
|
Domain Architecture |
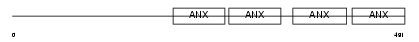
|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50) (56 kDa autoantigen). | |||||
|
CPSF4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 15) | NC score | 0.230331 (rank : 7) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
ZC11A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.128686 (rank : 15) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75152, Q6AHY4, Q6AHY9, Q6AW79, Q6AWA1, Q6PJK4, Q86XZ7 | Gene names | ZC3H11A, KIAA0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
TTP_MOUSE
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.105679 (rank : 20) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
CCNK_MOUSE
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.027434 (rank : 35) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
SIX5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.030345 (rank : 29) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 386 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N196 | Gene names | SIX5, DMAHP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein SIX5 (DM locus-associated homeodomain protein). | |||||
|
CPSF4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.209095 (rank : 8) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
TTP_HUMAN
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.100707 (rank : 21) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26651 | Gene names | ZFP36, G0S24, TIS11A, TTP | |||
|
Domain Architecture |
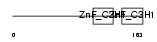
|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36 homolog) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (G0/G1 switch regulatory protein 24). | |||||
|
ZN207_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.066066 (rank : 26) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
ANX11_MOUSE
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.020561 (rank : 45) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
IF35_MOUSE
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.026457 (rank : 36) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
SIX5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.028898 (rank : 31) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70178 | Gene names | Six5, Dmahp | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX5 (DM locus-associated homeodomain protein homolog). | |||||
|
ZC11A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.106295 (rank : 19) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6NZF1, Q6NXW9, Q6PDR6, Q80TU7, Q8C5L5, Q99JN6 | Gene names | Zc3h11a, Kiaa0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
MKRN4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.095909 (rank : 23) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
PHF3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.026115 (rank : 37) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
DVL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.012962 (rank : 53) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
FMN2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.029062 (rank : 30) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
MBD6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.022382 (rank : 44) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
ZC3H8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.124695 (rank : 16) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
FHOD1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.024991 (rank : 40) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
CPSF7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.028550 (rank : 33) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N684, Q7Z3H9, Q9H025, Q9H9V1 | Gene names | CPSF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7 (Cleavage and polyadenylation specificity factor 59 kDa subunit) (CPSF 59 kDa subunit) (Pre-mRNA cleavage factor Im 59 kDa subunit). | |||||
|
CPSF7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.028509 (rank : 34) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BTV2, Q3TNF1, Q8BKE7, Q8CFS8 | Gene names | Cpsf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7. | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.016335 (rank : 48) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
MNT_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.025986 (rank : 38) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.023731 (rank : 43) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
RIPK3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.001740 (rank : 63) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QZL0 | Gene names | Ripk3, Rip3 | |||
|
Domain Architecture |
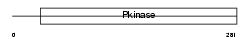
|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3) (mRIP3). | |||||
|
WASF2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.012048 (rank : 56) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.024432 (rank : 42) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.024713 (rank : 41) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
GORS2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.014424 (rank : 52) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8Y8, Q96I74, Q96K84, Q9H946, Q9UFW4 | Gene names | GORASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55) (p59) (Golgi phosphoprotein 6) (GOLPH6). | |||||
|
RBM12_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.019858 (rank : 46) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
SF3B4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.007253 (rank : 60) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
DIAP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.014611 (rank : 51) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
ESX1L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.007767 (rank : 59) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N693, Q7Z6K7 | Gene names | ESX1L, ESX1R | |||
|
Domain Architecture |

|
|||||
| Description | Extraembryonic, spermatogenesis, homeobox 1-like protein. | |||||
|
RBM12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.018873 (rank : 47) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
SP5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.001744 (rank : 62) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
SP5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.001494 (rank : 64) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.011560 (rank : 57) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ZC3H8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.120532 (rank : 17) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
BIRC6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.009759 (rank : 58) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NR09, Q9ULD1 | Gene names | BIRC6, KIAA1289 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 6 (Ubiquitin-conjugating BIR-domain enzyme apollon). | |||||
|
DHX57_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.028807 (rank : 32) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
EDA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.012452 (rank : 54) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92838, O75910, Q5JUM7, Q9UP77, Q9Y6L0, Q9Y6L1, Q9Y6L2, Q9Y6L3, Q9Y6L4 | Gene names | EDA, ED1 | |||
|
Domain Architecture |
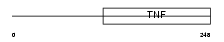
|
|||||
| Description | Ectodysplasin-A (Ectodermal dysplasia protein) (EDA protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
EDA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.012350 (rank : 55) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54693, O35705, Q9QWJ8, Q9QZ01, Q9QZ02 | Gene names | Eda, Ed1, Ta | |||
|
Domain Architecture |

|
|||||
| Description | Ectodysplasin-A (EDA protein homolog) (Tabby protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
MBL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.005529 (rank : 61) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41317 | Gene names | Mbl2 | |||
|
Domain Architecture |
|
|||||
| Description | Mannose-binding protein C precursor (MBP-C) (Mannan-binding protein) (RA-reactive factor P28A subunit) (RARF/P28A). | |||||
|
RBM22_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.032118 (rank : 27) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NW64, O95607 | Gene names | RBM22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
RBM22_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.032118 (rank : 28) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BHS3, Q3TJB0, Q9CXA0 | Gene names | Rbm22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.015142 (rank : 49) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.014711 (rank : 50) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
MKRN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.094153 (rank : 25) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
MKRN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.094516 (rank : 24) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
MKRN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.097763 (rank : 22) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
MBN3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 7.22167e-183 (rank : 1) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NUK0, Q5JXN8, Q5JXN9, Q5JXP4, Q6UDQ1, Q8TAD9, Q8TAF4, Q9H0Z7, Q9UF37 | Gene names | MBNL3, CHCR, MBLX39, MBXL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein HCHCR). | |||||
|
MBN3_MOUSE
|
||||||
| NC score | 0.989341 (rank : 2) | θ value | 3.25673e-167 (rank : 2) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8R003 | Gene names | Mbnl3, Chcr, Mbxl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein MCHCR). | |||||
|
MBNL_MOUSE
|
||||||
| NC score | 0.976171 (rank : 3) | θ value | 1.02895e-112 (rank : 3) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JKP5 | Gene names | Mbnl1, Exp, Mbnl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
MBNL_HUMAN
|
||||||
| NC score | 0.975617 (rank : 4) | θ value | 6.0463e-105 (rank : 4) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NR56, O43311, O43797 | Gene names | MBNL1, EXP, KIAA0428, MBNL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
ZC3H3_HUMAN
|
||||||
| NC score | 0.327497 (rank : 5) | θ value | 8.40245e-06 (rank : 6) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ZC3H3_MOUSE
|
||||||
| NC score | 0.326488 (rank : 6) | θ value | 6.43352e-06 (rank : 5) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
CPSF4_HUMAN
|
||||||
| NC score | 0.230331 (rank : 7) | θ value | 0.279714 (rank : 15) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
CPSF4_MOUSE
|
||||||
| NC score | 0.209095 (rank : 8) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
MKRN1_MOUSE
|
||||||
| NC score | 0.157751 (rank : 9) | θ value | 0.0148317 (rank : 7) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.150930 (rank : 10) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ZC3H6_HUMAN
|
||||||
| NC score | 0.149599 (rank : 11) | θ value | 0.0961366 (rank : 9) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
MKRN1_HUMAN
|
||||||
| NC score | 0.140773 (rank : 12) | θ value | 0.0961366 (rank : 8) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.138158 (rank : 13) | θ value | 0.21417 (rank : 12) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.136264 (rank : 14) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
ZC11A_HUMAN
|
||||||
| NC score | 0.128686 (rank : 15) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75152, Q6AHY4, Q6AHY9, Q6AW79, Q6AWA1, Q6PJK4, Q86XZ7 | Gene names | ZC3H11A, KIAA0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
ZC3H8_HUMAN
|
||||||
| NC score | 0.124695 (rank : 16) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
ZC3H8_MOUSE
|
||||||
| NC score | 0.120532 (rank : 17) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
MKRN3_HUMAN
|
||||||
| NC score | 0.120387 (rank : 18) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
ZC11A_MOUSE
|
||||||
| NC score | 0.106295 (rank : 19) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6NZF1, Q6NXW9, Q6PDR6, Q80TU7, Q8C5L5, Q99JN6 | Gene names | Zc3h11a, Kiaa0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
TTP_MOUSE
|
||||||
| NC score | 0.105679 (rank : 20) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
TTP_HUMAN
|
||||||
| NC score | 0.100707 (rank : 21) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26651 | Gene names | ZFP36, G0S24, TIS11A, TTP | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36 homolog) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (G0/G1 switch regulatory protein 24). | |||||
|
MKRN3_MOUSE
|
||||||
| NC score | 0.097763 (rank : 22) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
MKRN4_HUMAN
|
||||||
| NC score | 0.095909 (rank : 23) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
MKRN2_MOUSE
|
||||||
| NC score | 0.094516 (rank : 24) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
MKRN2_HUMAN
|
||||||
| NC score | 0.094153 (rank : 25) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.066066 (rank : 26) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
RBM22_HUMAN
|
||||||
| NC score | 0.032118 (rank : 27) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NW64, O95607 | Gene names | RBM22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
RBM22_MOUSE
|
||||||
| NC score | 0.032118 (rank : 28) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BHS3, Q3TJB0, Q9CXA0 | Gene names | Rbm22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
SIX5_HUMAN
|
||||||
| NC score | 0.030345 (rank : 29) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 386 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N196 | Gene names | SIX5, DMAHP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein SIX5 (DM locus-associated homeodomain protein). | |||||
|
FMN2_MOUSE
|
||||||
| NC score | 0.029062 (rank : 30) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
SIX5_MOUSE
|
||||||
| NC score | 0.028898 (rank : 31) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70178 | Gene names | Six5, Dmahp | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX5 (DM locus-associated homeodomain protein homolog). | |||||
|
DHX57_HUMAN
|
||||||
| NC score | 0.028807 (rank : 32) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
CPSF7_HUMAN
|
||||||
| NC score | 0.028550 (rank : 33) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N684, Q7Z3H9, Q9H025, Q9H9V1 | Gene names | CPSF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7 (Cleavage and polyadenylation specificity factor 59 kDa subunit) (CPSF 59 kDa subunit) (Pre-mRNA cleavage factor Im 59 kDa subunit). | |||||
|
CPSF7_MOUSE
|
||||||
| NC score | 0.028509 (rank : 34) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BTV2, Q3TNF1, Q8BKE7, Q8CFS8 | Gene names | Cpsf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7. | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.027434 (rank : 35) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
IF35_MOUSE
|
||||||
| NC score | 0.026457 (rank : 36) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
PHF3_HUMAN
|
||||||
| NC score | 0.026115 (rank : 37) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.025986 (rank : 38) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
ANX11_HUMAN
|
||||||
| NC score | 0.025930 (rank : 39) | θ value | 0.279714 (rank : 14) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50995 | Gene names | ANXA11, ANX11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50) (56 kDa autoantigen). | |||||
|
FHOD1_MOUSE
|
||||||
| NC score | 0.024991 (rank : 40) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.024713 (rank : 41) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.024432 (rank : 42) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.023731 (rank : 43) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.022382 (rank : 44) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
ANX11_MOUSE
|
||||||
| NC score | 0.020561 (rank : 45) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
RBM12_MOUSE
|
||||||
| NC score | 0.019858 (rank : 46) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
RBM12_HUMAN
|
||||||
| NC score | 0.018873 (rank : 47) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.016335 (rank : 48) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.015142 (rank : 49) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.014711 (rank : 50) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
DIAP1_MOUSE
|
||||||
| NC score | 0.014611 (rank : 51) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
GORS2_HUMAN
|
||||||
| NC score | 0.014424 (rank : 52) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8Y8, Q96I74, Q96K84, Q9H946, Q9UFW4 | Gene names | GORASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55) (p59) (Golgi phosphoprotein 6) (GOLPH6). | |||||
|
DVL2_HUMAN
|
||||||
| NC score | 0.012962 (rank : 53) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
EDA_HUMAN
|
||||||
| NC score | 0.012452 (rank : 54) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92838, O75910, Q5JUM7, Q9UP77, Q9Y6L0, Q9Y6L1, Q9Y6L2, Q9Y6L3, Q9Y6L4 | Gene names | EDA, ED1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectodysplasin-A (Ectodermal dysplasia protein) (EDA protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
EDA_MOUSE
|
||||||
| NC score | 0.012350 (rank : 55) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54693, O35705, Q9QWJ8, Q9QZ01, Q9QZ02 | Gene names | Eda, Ed1, Ta | |||
|
Domain Architecture |

|
|||||
| Description | Ectodysplasin-A (EDA protein homolog) (Tabby protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
WASF2_MOUSE
|
||||||
| NC score | 0.012048 (rank : 56) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.011560 (rank : 57) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
BIRC6_HUMAN
|
||||||
| NC score | 0.009759 (rank : 58) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NR09, Q9ULD1 | Gene names | BIRC6, KIAA1289 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 6 (Ubiquitin-conjugating BIR-domain enzyme apollon). | |||||
|
ESX1L_HUMAN
|
||||||
| NC score | 0.007767 (rank : 59) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N693, Q7Z6K7 | Gene names | ESX1L, ESX1R | |||
|
Domain Architecture |

|
|||||
| Description | Extraembryonic, spermatogenesis, homeobox 1-like protein. | |||||
|
SF3B4_HUMAN
|
||||||
| NC score | 0.007253 (rank : 60) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
MBL2_MOUSE
|
||||||
| NC score | 0.005529 (rank : 61) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41317 | Gene names | Mbl2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannose-binding protein C precursor (MBP-C) (Mannan-binding protein) (RA-reactive factor P28A subunit) (RARF/P28A). | |||||
|
SP5_HUMAN
|
||||||
| NC score | 0.001744 (rank : 62) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
RIPK3_MOUSE
|
||||||
| NC score | 0.001740 (rank : 63) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QZL0 | Gene names | Ripk3, Rip3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3) (mRIP3). | |||||
|
SP5_MOUSE
|
||||||
| NC score | 0.001494 (rank : 64) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||