Please be patient as the page loads
|
BIRC6_HUMAN
|
||||||
| SwissProt Accessions | Q9NR09, Q9ULD1 | Gene names | BIRC6, KIAA1289 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 6 (Ubiquitin-conjugating BIR-domain enzyme apollon). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
BIRC6_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 161 | |
| SwissProt Accessions | Q9NR09, Q9ULD1 | Gene names | BIRC6, KIAA1289 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 6 (Ubiquitin-conjugating BIR-domain enzyme apollon). | |||||
|
BIRC4_HUMAN
|
||||||
| θ value | 2.0648e-12 (rank : 2) | NC score | 0.410027 (rank : 53) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P98170, Q9NQ14 | Gene names | BIRC4, API3, IAP3, XIAP | |||
|
Domain Architecture |
|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP-like protein) (HILP). | |||||
|
BIRC4_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 3) | NC score | 0.409424 (rank : 54) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60989, O08865 | Gene names | Birc4, Aipa, Api3, Miha, Xiap | |||
|
Domain Architecture |
|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP homolog A) (MIAP3) (MIAP-3). | |||||
|
UBE2A_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 4) | NC score | 0.606817 (rank : 4) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P49459, Q4TTG1, Q96FX4 | Gene names | UBE2A, RAD6A | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 A (EC 6.3.2.19) (Ubiquitin-protein ligase A) (Ubiquitin carrier protein A) (HR6A) (hHR6A). | |||||
|
UBE2A_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 5) | NC score | 0.606817 (rank : 5) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Z255 | Gene names | Ube2a, Rad6a | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 A (EC 6.3.2.19) (Ubiquitin-protein ligase A) (Ubiquitin carrier protein A) (HR6A) (mHR6A). | |||||
|
BIRC5_MOUSE
|
||||||
| θ value | 1.33837e-11 (rank : 6) | NC score | 0.478743 (rank : 49) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70201, Q923F7, Q9WU53, Q9WU54 | Gene names | Birc5, Api4, Iap4 | |||
|
Domain Architecture |
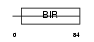
|
|||||
| Description | Baculoviral IAP repeat-containing protein 5 (Apoptosis inhibitor survivin) (Apoptosis inhibitor 4) (TIAP). | |||||
|
UBE2B_HUMAN
|
||||||
| θ value | 1.74796e-11 (rank : 7) | NC score | 0.605314 (rank : 6) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P63146, P23567, Q4PJ15, Q9D0J6 | Gene names | UBE2B, RAD6B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 B (EC 6.3.2.19) (Ubiquitin-protein ligase B) (Ubiquitin carrier protein B) (HR6B) (hHR6B) (E2-17 kDa). | |||||
|
UBE2B_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 8) | NC score | 0.605314 (rank : 7) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P63147, P23567, Q3UGS2, Q9D0J6 | Gene names | Ube2b, Rad6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 B (EC 6.3.2.19) (Ubiquitin-protein ligase B) (Ubiquitin carrier protein B) (HR6B) (E214K). | |||||
|
BIRC5_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 9) | NC score | 0.473040 (rank : 50) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15392, Q2I3N8, Q4VGX0, Q53F61, Q5MGC6, Q6FHL2, Q75SP2, Q9P2W8 | Gene names | BIRC5, API4, IAP4 | |||
|
Domain Architecture |
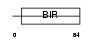
|
|||||
| Description | Baculoviral IAP repeat-containing protein 5 (Apoptosis inhibitor survivin) (Apoptosis inhibitor 4). | |||||
|
UBE2T_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 10) | NC score | 0.596853 (rank : 14) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NPD8 | Gene names | UBE2T | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 T (EC 6.3.2.19) (Ubiquitin-protein ligase T) (Ubiquitin carrier protein T). | |||||
|
BIRC2_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 11) | NC score | 0.380637 (rank : 56) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13490, Q16516, Q4TTG0 | Gene names | BIRC2, API1, IAP2, MIHB | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (HIAP2) (HIAP-2) (C-IAP1) (TNFR2-TRAF signaling complex protein 2) (IAP homolog B). | |||||
|
BIRC3_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 12) | NC score | 0.380896 (rank : 55) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13489, Q16628, Q9HC27, Q9UP46 | Gene names | BIRC3, API2, IAP1, MIHC | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (HIAP1) (HIAP-1) (C-IAP2) (TNFR2-TRAF signaling complex protein 1) (IAP homolog C) (Apoptosis inhibitor 2) (API2). | |||||
|
UB2G2_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 13) | NC score | 0.608468 (rank : 2) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P60604, P56554 | Gene names | UBE2G2 | |||
|
Domain Architecture |
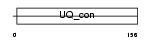
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 G2 (EC 6.3.2.19) (Ubiquitin-protein ligase G2) (Ubiquitin carrier protein G2). | |||||
|
UB2G2_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 14) | NC score | 0.608468 (rank : 3) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P60605, P56554 | Gene names | Ube2g2 | |||
|
Domain Architecture |
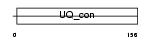
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 G2 (EC 6.3.2.19) (Ubiquitin-protein ligase G2) (Ubiquitin carrier protein G2). | |||||
|
BIRC2_MOUSE
|
||||||
| θ value | 1.63604e-09 (rank : 15) | NC score | 0.379131 (rank : 57) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62210, O08864 | Gene names | Birc2, Birc3, Iap2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (MIAP2) (MIAP-2). | |||||
|
UB2R1_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 16) | NC score | 0.598606 (rank : 11) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P49427 | Gene names | CDC34, UBE2R1 | |||
|
Domain Architecture |
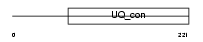
|
|||||
| Description | Ubiquitin-conjugating enzyme E2-32 kDa complementing (EC 6.3.2.19) (Ubiquitin-protein ligase) (Ubiquitin carrier protein) (E2-CDC34). | |||||
|
UB2R1_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 17) | NC score | 0.598701 (rank : 10) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8CFI2, Q505K8 | Gene names | Cdc34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2-32 kDa complementing (EC 6.3.2.19) (Ubiquitin-protein ligase) (Ubiquitin carrier protein) (E2-CDC34). | |||||
|
UB2G1_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 18) | NC score | 0.597347 (rank : 12) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P62253, Q99462 | Gene names | UBE2G1, UBE2G | |||
|
Domain Architecture |
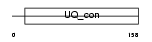
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 G1 (EC 6.3.2.19) (Ubiquitin-protein ligase G1) (Ubiquitin carrier protein G1) (E217K) (UBC7). | |||||
|
UB2G1_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 19) | NC score | 0.597347 (rank : 13) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P62254, Q99462 | Gene names | Ube2g1, Ube2g | |||
|
Domain Architecture |
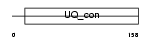
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 G1 (EC 6.3.2.19) (Ubiquitin-protein ligase G1) (Ubiquitin carrier protein G1) (E217K) (UBC7). | |||||
|
UBE2T_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 20) | NC score | 0.590668 (rank : 17) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9CQ37 | Gene names | Ube2t | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 T (EC 6.3.2.19) (Ubiquitin-protein ligase T) (Ubiquitin carrier protein T). | |||||
|
BIRC3_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 21) | NC score | 0.378831 (rank : 58) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08863 | Gene names | Birc3, Birc2, Iap1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (MIAP1) (MIAP-1). | |||||
|
UBC9_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 22) | NC score | 0.601797 (rank : 8) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P63279, P50550, Q15698, Q86VB3 | Gene names | UBE2I, UBC9, UBCE9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SUMO-1-conjugating enzyme UBC9 (EC 6.3.2.19) (SUMO-1-protein ligase) (Ubiquitin-conjugating enzyme E2 I) (Ubiquitin-protein ligase I) (Ubiquitin carrier protein I) (Ubiquitin carrier protein 9) (p18). | |||||
|
UBC9_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 23) | NC score | 0.601797 (rank : 9) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P63280, P50550, Q15698, Q86VB3 | Gene names | Ube2i, Ubc9, Ubce9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SUMO-1-conjugating enzyme UBC9 (EC 6.3.2.19) (SUMO-1-protein ligase) (Ubiquitin-conjugating enzyme E2 I) (Ubiquitin-protein ligase I) (Ubiquitin carrier protein I) (Ubiquitin carrier protein 9) (mUBC9). | |||||
|
UBE2N_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 24) | NC score | 0.594026 (rank : 15) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P61088, Q16781 | Gene names | UBE2N, BLU | |||
|
Domain Architecture |
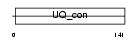
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 N (EC 6.3.2.19) (Ubiquitin-protein ligase N) (Ubiquitin carrier protein N) (Ubc13) (Bendless-like ubiquitin-conjugating enzyme). | |||||
|
UBE2N_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 25) | NC score | 0.594026 (rank : 16) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P61089, Q16781, Q6ZWZ0, Q9DAJ6 | Gene names | Ube2n, Blu | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 N (EC 6.3.2.19) (Ubiquitin-protein ligase N) (Ubiquitin carrier protein N) (Ubc13) (Bendless-like ubiquitin-conjugating enzyme). | |||||
|
BIR1B_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 26) | NC score | 0.332930 (rank : 61) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QUK4, O09124, Q9R030 | Gene names | Birc1b, Naip-rs6, Naip2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1b (Neuronal apoptosis inhibitory protein 2). | |||||
|
UB2D2_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 27) | NC score | 0.575811 (rank : 21) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P62837, P51669 | Gene names | UBE2D2, UBC4, UBCH5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 D2 (EC 6.3.2.19) (Ubiquitin-protein ligase D2) (Ubiquitin carrier protein D2) (Ubiquitin-conjugating enzyme E2-17 kDa 2) (E2(17)KB 2). | |||||
|
UB2D2_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 28) | NC score | 0.575811 (rank : 22) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P62838, P51669 | Gene names | Ube2d2, Ubc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 D2 (EC 6.3.2.19) (Ubiquitin-protein ligase D2) (Ubiquitin carrier protein D2) (Ubiquitin-conjugating enzyme E2-17 kDa 2) (E2(17)KB 2). | |||||
|
UB2D3_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 29) | NC score | 0.575688 (rank : 23) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P61077, P47986, Q6IB88, Q6NXS4 | Gene names | UBE2D3, UBCH5C | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 D3 (EC 6.3.2.19) (Ubiquitin-protein ligase D3) (Ubiquitin carrier protein D3) (Ubiquitin-conjugating enzyme E2-17 kDa 3) (E2(17)KB 3). | |||||
|
UB2D3_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 30) | NC score | 0.575688 (rank : 24) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P61079 | Gene names | Ube2d3 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 D3 (EC 6.3.2.19) (Ubiquitin-protein ligase D3) (Ubiquitin carrier protein D3) (Ubiquitin-conjugating enzyme E2-17 kDa 3) (E2(17)KB 3). | |||||
|
UB2L6_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 31) | NC score | 0.585967 (rank : 20) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9QZU9, Q922F1, Q9CQN0 | Gene names | Ube2l6, Ubce8 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 L6 (EC 6.3.2.19) (Ubiquitin-protein ligase L6) (Ubiquitin carrier protein L6) (UbcM8). | |||||
|
BIRC7_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 32) | NC score | 0.354464 (rank : 60) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96CA5, Q9BQV0, Q9H2A8, Q9HAP7 | Gene names | BIRC7, KIAP, LIVIN, MLIAP | |||
|
Domain Architecture |
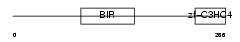
|
|||||
| Description | Baculoviral IAP repeat-containing protein 7 (Kidney inhibitor of apoptosis protein) (KIAP) (Melanoma inhibitor of apoptosis protein) (ML-IAP) (Livin). | |||||
|
UBE2S_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 33) | NC score | 0.587950 (rank : 19) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16763, Q9BTC1 | Gene names | UBE2S, E2EPF | |||
|
Domain Architecture |
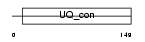
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 S (EC 6.3.2.19) (Ubiquitin-protein ligase S) (Ubiquitin carrier protein S) (Ubiquitin-conjugating enzyme E2-24 kDa) (E2-EPF5). | |||||
|
UBE2S_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 34) | NC score | 0.588078 (rank : 18) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q921J4, Q3U4V7 | Gene names | Ube2s, E2epf | |||
|
Domain Architecture |
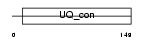
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 S (EC 6.3.2.19) (Ubiquitin-protein ligase S) (Ubiquitin carrier protein S) (Ubiquitin-conjugating enzyme E2-24 kDa) (E2-EPF5). | |||||
|
UB2D1_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 35) | NC score | 0.572369 (rank : 27) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P51668 | Gene names | UBE2D1, UBCH5, UBCH5A | |||
|
Domain Architecture |
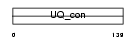
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 D1 (EC 6.3.2.19) (Ubiquitin-protein ligase D1) (Ubiquitin carrier protein D1) (UbcH5) (Ubiquitin- conjugating enzyme E2-17 kDa 1) (E2(17)KB 1). | |||||
|
UB2D1_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 36) | NC score | 0.572369 (rank : 28) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P61080 | Gene names | Ube2d1 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 D1 (EC 6.3.2.19) (Ubiquitin-protein ligase D1) (Ubiquitin carrier protein D1) (Ubiquitin-conjugating enzyme E2-17 kDa 1) (E2(17)KB 1). | |||||
|
BIRC1_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 37) | NC score | 0.331518 (rank : 62) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13075, O75857, Q13730, Q99796 | Gene names | BIRC1, NAIP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1 (Neuronal apoptosis inhibitory protein). | |||||
|
BIR1F_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 38) | NC score | 0.325611 (rank : 64) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JIB6, O09121, O09122, P81704 | Gene names | Birc1f, Naip-rs4, Naip6 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1f (Neuronal apoptosis inhibitory protein 6). | |||||
|
BIR1A_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 39) | NC score | 0.324270 (rank : 66) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QWK5, Q9JIB5, Q9R017 | Gene names | Birc1a, Naip, Naip1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1a (Neuronal apoptosis inhibitory protein 1). | |||||
|
BIR1E_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 40) | NC score | 0.325691 (rank : 63) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9R016, O09121, O09122, P81703, Q9R029 | Gene names | Birc1e, Naip-rs3, Naip5 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1e (Neuronal apoptosis inhibitory protein 5). | |||||
|
BIR1G_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 41) | NC score | 0.325202 (rank : 65) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JIB3 | Gene names | Birc1g, Naip7 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1g (Neuronal apoptosis inhibitory protein 7). | |||||
|
BIRC8_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 42) | NC score | 0.371664 (rank : 59) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96P09, Q96RW5 | Gene names | BIRC8, ILP2 | |||
|
Domain Architecture |
|
|||||
| Description | Baculoviral IAP repeat-containing protein 8 (Inhibitor of apoptosis- like protein 2) (IAP-like protein 2) (ILP-2) (Testis-specific inhibitor of apoptosis). | |||||
|
UB2L3_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 43) | NC score | 0.575233 (rank : 25) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P68036, P51966, P70653, Q9HAV1 | Gene names | UBE2L3, UBCE7, UBCH7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 L3 (EC 6.3.2.19) (Ubiquitin-protein ligase L3) (Ubiquitin carrier protein L3) (UbcH7) (E2-F1) (L-UBC). | |||||
|
UB2L3_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 44) | NC score | 0.575233 (rank : 26) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P68037, P51966, P70653, Q9HAV1 | Gene names | Ube2l3, Ubce7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 L3 (EC 6.3.2.19) (Ubiquitin-protein ligase L3) (Ubiquitin carrier protein L3) (UbcM4). | |||||
|
UBE2C_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 45) | NC score | 0.568887 (rank : 30) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O00762 | Gene names | UBE2C, UBCH10 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 C (EC 6.3.2.19) (Ubiquitin-protein ligase C) (Ubiquitin carrier protein C) (UbcH10). | |||||
|
UB2L6_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 46) | NC score | 0.569208 (rank : 29) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O14933, Q9UEZ0 | Gene names | UBE2L6, UBCH8 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 L6 (EC 6.3.2.19) (Ubiquitin-protein ligase L6) (Ubiquitin carrier protein L6) (UbcH8) (Retinoic acid- induced gene B protein) (RIG-B). | |||||
|
UB2E3_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 47) | NC score | 0.551164 (rank : 34) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q969T4, Q5U0R7, Q7Z4W4 | Gene names | UBE2E3, UBCE4, UBCH9 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 E3 (EC 6.3.2.19) (Ubiquitin-protein ligase E3) (Ubiquitin carrier protein E3) (Ubiquitin-conjugating enzyme E2-23 kDa) (UbcH9) (UbcM2). | |||||
|
UB2E3_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 48) | NC score | 0.551164 (rank : 35) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P52483, O09180, Q3TI00, Q91X63 | Gene names | Ube2e3, Ubce4, Ubcm2 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 E3 (EC 6.3.2.19) (Ubiquitin-protein ligase E3) (Ubiquitin carrier protein E3) (Ubiquitin-conjugating enzyme E2-23 kDa) (UbcM2). | |||||
|
UB2E2_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 49) | NC score | 0.549768 (rank : 36) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96LR5 | Gene names | UBE2E2, UBCH8 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 E2 (EC 6.3.2.19) (Ubiquitin-protein ligase E2) (Ubiquitin carrier protein E2) (UbcH8). | |||||
|
UB2E2_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 50) | NC score | 0.548978 (rank : 37) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q91W82 | Gene names | Ube2e2 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 E2 (EC 6.3.2.19) (Ubiquitin-protein ligase E2) (Ubiquitin carrier protein E2). | |||||
|
UBC1_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 51) | NC score | 0.554990 (rank : 32) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P61086, O54806, P27924, Q16721, Q9CVV9, Q9Y2D3 | Gene names | HIP2, LIG | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2-25 kDa (EC 6.3.2.19) (Ubiquitin- protein ligase) (Ubiquitin carrier protein) (E2(25K)) (Huntingtin- interacting protein 2) (HIP-2). | |||||
|
UBC1_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 52) | NC score | 0.554990 (rank : 33) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P61087, O54806, P27924, Q16721, Q9CVV9 | Gene names | Hip2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2-25 kDa (EC 6.3.2.19) (Ubiquitin- protein ligase) (Ubiquitin carrier protein) (E2(25K)) (Huntingtin- interacting protein 2) (HIP-2). | |||||
|
UBE2C_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 53) | NC score | 0.564860 (rank : 31) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9D1C1, Q3TUJ0 | Gene names | Ube2c, Ubch10 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 C (EC 6.3.2.19) (Ubiquitin-protein ligase C) (Ubiquitin carrier protein C) (UbcH10). | |||||
|
UB2E1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 54) | NC score | 0.547314 (rank : 38) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P51965 | Gene names | UBE2E1, UBCH6 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 E1 (EC 6.3.2.19) (Ubiquitin-protein ligase E1) (Ubiquitin carrier protein E1) (UbcH6). | |||||
|
UB2E1_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 55) | NC score | 0.545927 (rank : 39) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P52482 | Gene names | Ube2e1, Ubce5, Ubcm3 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 E1 (EC 6.3.2.19) (Ubiquitin-protein ligase E1) (Ubiquitin carrier protein E1) (UbcM3). | |||||
|
UBC12_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 56) | NC score | 0.542311 (rank : 40) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P61081, O76069, Q8VC50 | Gene names | UBE2M, UBC12 | |||
|
Domain Architecture |
|
|||||
| Description | NEDD8-conjugating enzyme Ubc12 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme E2 M) (NEDD8 protein ligase) (NEDD8 carrier protein). | |||||
|
UBC12_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 57) | NC score | 0.542311 (rank : 41) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P61082, O76069, Q8VC50 | Gene names | Ube2m, Ubc-rs2, Ubc12 | |||
|
Domain Architecture |
|
|||||
| Description | NEDD8-conjugating enzyme Ubc12 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme E2 M) (NEDD8 protein ligase) (NEDD8 carrier protein). | |||||
|
UB2J2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 58) | NC score | 0.486000 (rank : 48) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8N2K1, Q96N26, Q96T84 | Gene names | UBE2J2, NCUBE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 J2 (EC 6.3.2.19) (Non-canonical ubiquitin-conjugating enzyme 2) (NCUBE2). | |||||
|
UB2J2_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 59) | NC score | 0.488205 (rank : 47) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6P073, Q8C6A1, Q91Y64, Q9CWY5, Q9DC18, Q9QX58 | Gene names | Ube2j2, Ncube2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 J2 (EC 6.3.2.19) (Non-canonical ubiquitin-conjugating enzyme 2) (NCUBE2). | |||||
|
UB2J1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 60) | NC score | 0.451044 (rank : 51) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y385, Q5W0N4, Q9BZ32, Q9NQL3, Q9NY66, Q9P011, Q9P0S0, Q9UF10 | Gene names | UBE2J1, NCUBE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 J1 (EC 6.3.2.19) (Non-canonical ubiquitin-conjugating enzyme 1) (NCUBE1) (Yeast ubiquitin-conjugating enzyme UBC6 homolog E) (HSUBC6e). | |||||
|
UB2J1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 61) | NC score | 0.450171 (rank : 52) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JJZ4, Q9DC92 | Gene names | Ube2j1, Ncube1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 J1 (EC 6.3.2.19) (Non-canonical ubiquitin-conjugating enzyme 1) (NCUBE1). | |||||
|
IFT74_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 62) | NC score | 0.032802 (rank : 90) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
UBE2H_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 63) | NC score | 0.533438 (rank : 42) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P62256, P37286 | Gene names | UBE2H | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 H (EC 6.3.2.19) (Ubiquitin-protein ligase H) (Ubiquitin carrier protein H) (UbcH2) (E2-20K). | |||||
|
UBE2H_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 64) | NC score | 0.533438 (rank : 43) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P62257, P37286 | Gene names | Ube2h | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 H (EC 6.3.2.19) (Ubiquitin-protein ligase H) (Ubiquitin carrier protein H) (UBCH2) (E2-20K). | |||||
|
UBE2W_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 65) | NC score | 0.511142 (rank : 45) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8VDW4, Q9D5H3 | Gene names | Ube2w | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin-conjugating enzyme E2 W (EC 6.3.2.19) (Ubiquitin- protein ligase W) (Ubiquitin carrier protein W). | |||||
|
UBE2W_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 66) | NC score | 0.509921 (rank : 46) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96B02, Q9H823, Q9HAG6, Q9NV07 | Gene names | UBE2W | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin-conjugating enzyme E2 W (EC 6.3.2.19) (Ubiquitin- protein ligase W) (Ubiquitin carrier protein W). | |||||
|
USP9Y_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 67) | NC score | 0.039410 (rank : 85) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
ARRS_MOUSE
|
||||||
| θ value | 0.279714 (rank : 68) | NC score | 0.034500 (rank : 89) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20443, Q91W62 | Gene names | Sag | |||
|
Domain Architecture |
|
|||||
| Description | S-arrestin (Retinal S-antigen) (48 kDa protein) (S-AG) (Rod photoreceptor arrestin). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 0.365318 (rank : 69) | NC score | 0.020920 (rank : 109) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
LRC45_HUMAN
|
||||||
| θ value | 0.365318 (rank : 70) | NC score | 0.022449 (rank : 105) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
FOXP4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 71) | NC score | 0.016191 (rank : 122) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IVH2, Q96E19 | Gene names | FOXP4, FKHLA | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P4 (Fork head-related protein-like A). | |||||
|
LRRF1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 72) | NC score | 0.031924 (rank : 91) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 73) | NC score | 0.016062 (rank : 124) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
DYH5_MOUSE
|
||||||
| θ value | 0.62314 (rank : 74) | NC score | 0.021898 (rank : 106) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
FEZ2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 75) | NC score | 0.035797 (rank : 88) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UHY8, Q76LN0, Q99690 | Gene names | FEZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 2 (Zygin-2) (Zygin II). | |||||
|
SCFD2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 76) | NC score | 0.039910 (rank : 83) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BTY8, Q3USN5, Q8BFV9, Q8BKU5, Q8BKZ0, Q8BTP6, Q8BTQ9, Q8BXI1, Q8BXS0, Q8BZ58 | Gene names | Scfd2, Stxbp1l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sec1 family domain-containing protein 2 (Syntaxin-binding protein 1- like 1) (Neuronal Sec1). | |||||
|
UBE2U_HUMAN
|
||||||
| θ value | 0.62314 (rank : 77) | NC score | 0.515188 (rank : 44) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5VVX9, Q8N1D4 | Gene names | UBE2U | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 U (EC 6.3.2.19) (Ubiquitin-protein ligase U) (Ubiquitin carrier protein U). | |||||
|
ANR18_HUMAN
|
||||||
| θ value | 0.813845 (rank : 78) | NC score | 0.014644 (rank : 132) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1106 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IVF6, Q7Z468 | Gene names | ANKRD18A, KIAA2015 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 18A. | |||||
|
CI093_HUMAN
|
||||||
| θ value | 0.813845 (rank : 79) | NC score | 0.018753 (rank : 115) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
P3H3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 80) | NC score | 0.023744 (rank : 103) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IVL6, Q13512, Q15740, Q66K32, Q6NX61, Q7L2T1 | Gene names | LEPREL2, P3H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B). | |||||
|
WDR75_HUMAN
|
||||||
| θ value | 0.813845 (rank : 81) | NC score | 0.036436 (rank : 87) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWA0, Q96J10, Q9H8U8, Q9H9U5, Q9H9V8, Q9UIX2 | Gene names | WDR75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 75. | |||||
|
CLN3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 82) | NC score | 0.038358 (rank : 86) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13286, O00668, Q9UP09, Q9UP11, Q9UP12, Q9UP13, Q9UP14 | Gene names | CLN3, BTS | |||
|
Domain Architecture |
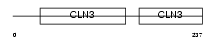
|
|||||
| Description | Protein CLN3 (Battenin) (Batten disease protein). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 83) | NC score | 0.014666 (rank : 131) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
K0423_HUMAN
|
||||||
| θ value | 1.06291 (rank : 84) | NC score | 0.028832 (rank : 95) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 1.06291 (rank : 85) | NC score | 0.009670 (rank : 151) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
PK3CB_HUMAN
|
||||||
| θ value | 1.06291 (rank : 86) | NC score | 0.019056 (rank : 113) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42338 | Gene names | PIK3CB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
TS101_HUMAN
|
||||||
| θ value | 1.06291 (rank : 87) | NC score | 0.039858 (rank : 84) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99816, Q9BUM5 | Gene names | TSG101 | |||
|
Domain Architecture |
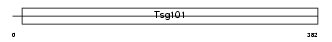
|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
VWF_MOUSE
|
||||||
| θ value | 1.06291 (rank : 88) | NC score | 0.012966 (rank : 140) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
CLSPN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 89) | NC score | 0.026876 (rank : 97) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
PLCB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 90) | NC score | 0.023766 (rank : 102) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NQ66, O60325, Q5TFF7, Q5TGC9, Q8IV93, Q9BQW2, Q9H4H2, Q9H8H5, Q9NQ65, Q9NQH9, Q9NTH4, Q9UJP6, Q9UM26 | Gene names | PLCB1, KIAA0581 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
SPTN5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 91) | NC score | 0.013786 (rank : 135) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
CCD62_HUMAN
|
||||||
| θ value | 1.81305 (rank : 92) | NC score | 0.023229 (rank : 104) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6P9F0, Q6ZVF2, Q86VJ0, Q9BYZ5 | Gene names | CCDC62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 60 (Aaa-protein) (Protein TSP- NY). | |||||
|
CRGB_HUMAN
|
||||||
| θ value | 1.81305 (rank : 93) | NC score | 0.013300 (rank : 138) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07316 | Gene names | CRYGB, CRYG2 | |||
|
Domain Architecture |
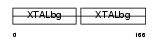
|
|||||
| Description | Gamma crystallin B (Gamma crystallin 1-2). | |||||
|
OX1R_HUMAN
|
||||||
| θ value | 1.81305 (rank : 94) | NC score | -0.000020 (rank : 175) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 909 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43613 | Gene names | HCRTR1 | |||
|
Domain Architecture |
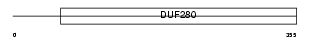
|
|||||
| Description | Orexin receptor type 1 (Ox1r) (Hypocretin receptor type 1). | |||||
|
PLCB1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 95) | NC score | 0.024984 (rank : 100) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z1B3, Q62075, Q8K5A5, Q8K5A6, Q9Z0E5, Q9Z2T5 | Gene names | Plcb1, Plcb | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
IFT74_MOUSE
|
||||||
| θ value | 2.36792 (rank : 96) | NC score | 0.025839 (rank : 98) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
RABE1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 97) | NC score | 0.017896 (rank : 116) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
TM108_HUMAN
|
||||||
| θ value | 2.36792 (rank : 98) | NC score | 0.025073 (rank : 99) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6UXF1, Q9BQH1, Q9BW81, Q9C0H3 | Gene names | TMEM108, KIAA1690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
USP9X_HUMAN
|
||||||
| θ value | 2.36792 (rank : 99) | NC score | 0.031056 (rank : 93) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
USP9X_MOUSE
|
||||||
| θ value | 2.36792 (rank : 100) | NC score | 0.031155 (rank : 92) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
WD51B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 101) | NC score | 0.007186 (rank : 159) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BHD1 | Gene names | Wdr51b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 51B. | |||||
|
GP176_MOUSE
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.005556 (rank : 166) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80WT4, Q80UC4 | Gene names | Gpr176, Agr9, Gm1012 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 176 (G-protein coupled receptor AGR9). | |||||
|
HPS5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.021531 (rank : 107) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPZ3, O95942, Q8N4U0 | Gene names | HPS5, AIBP63, KIAA1017 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 5 protein (Alpha-integrin-binding protein 63) (Ruby-eye protein 2 homolog) (Ru2). | |||||
|
P5CS_MOUSE
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.027896 (rank : 96) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z110, Q9R1P6 | Gene names | Aldh18a1, P5cs, Pycs | |||
|
Domain Architecture |

|
|||||
| Description | Delta 1-pyrroline-5-carboxylate synthetase (P5CS) (Aldehyde dehydrogenase 18 family member A1) [Includes: Glutamate 5-kinase (EC 2.7.2.11) (Gamma-glutamyl kinase) (GK); Gamma-glutamyl phosphate reductase (GPR) (EC 1.2.1.41) (Glutamate-5-semialdehyde dehydrogenase) (Glutamyl-gamma-semialdehyde dehydrogenase)]. | |||||
|
PKD1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 105) | NC score | 0.011053 (rank : 147) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.013580 (rank : 136) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
SFRP5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.017273 (rank : 119) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T4F7, O14780, Q86TH7 | Gene names | SFRP5, FRP1B, SARP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 5 precursor (sFRP-5) (Secreted apoptosis-related protein 3) (SARP-3) (Frizzled-related protein 1b) (FRP-1b). | |||||
|
WD51B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.006631 (rank : 161) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TC44 | Gene names | WDR51B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 51B. | |||||
|
ARRS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.023851 (rank : 101) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10523, Q53SV3, Q99858 | Gene names | SAG | |||
|
Domain Architecture |
|
|||||
| Description | S-arrestin (Retinal S-antigen) (48 kDa protein) (S-AG) (Rod photoreceptor arrestin). | |||||
|
FAS_MOUSE
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.014881 (rank : 130) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19096, Q6PB72, Q8C4Z0, Q9EQR0 | Gene names | Fasn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
K1C14_MOUSE
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.006409 (rank : 163) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61781, Q91VQ4 | Gene names | Krt14, Krt1-14 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 14 (Cytokeratin-14) (CK-14) (Keratin-14) (K14). | |||||
|
PK3CB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.015391 (rank : 128) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BTI9 | Gene names | Pik3cb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PXDC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.016641 (rank : 121) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91ZV7, Q8BM20, Q9CWV5 | Gene names | Plxdc1, Tem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 1 precursor (Tumor endothelial marker 7). | |||||
|
RCOR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.017623 (rank : 117) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKL0, Q15044, Q6P2I9, Q86VG5 | Gene names | RCOR1, KIAA0071, RCOR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.017437 (rank : 118) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CFE3, Q3V092, Q8BK28, Q8CHI2 | Gene names | Rcor1, D12Wsu95e, Kiaa0071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
S35B2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.019651 (rank : 111) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TB61, Q5T9W1, Q5T9W2, Q7Z2G3, Q8NBK6, Q96AR6 | Gene names | SLC35B2, PAPST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenosine 3'-phospho 5'-phosphosulfate transporter 1 (PAPS transporter 1) (Solute carrier family 35 member B2) (Putative MAPK-activating protein PM15) (Putative NF-kappa-B-activating protein 48). | |||||
|
TNR1B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.013041 (rank : 139) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P25119, O88734, P97893 | Gene names | Tnfrsf1b, Tnfr-2, Tnfr2 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor). | |||||
|
TRAIP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 118) | NC score | 0.015653 (rank : 126) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
ABCAD_MOUSE
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.005167 (rank : 168) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SSE9, Q80T20, Q8BHZ2, Q8CB91 | Gene names | Abca13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
ANR38_MOUSE
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.011000 (rank : 148) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 512 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P9J5, Q8BV03 | Gene names | Ankrd38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 38. | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.011526 (rank : 146) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
FA45B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.016190 (rank : 123) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6NSW5, Q9NZ36 | Gene names | FAM45B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM45B. | |||||
|
GAK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | -0.002963 (rank : 176) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
LARGE_MOUSE
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.011970 (rank : 144) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1M7, Q497S9, Q6P7U2, Q80TW0 | Gene names | Large, Kiaa0609, Large1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycosyltransferase-like protein LARGE1 (EC 2.4.-.-) (Acetylglucosaminyltransferase-like 1A). | |||||
|
LRC19_HUMAN
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.006561 (rank : 162) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H756 | Gene names | LRRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 19 precursor. | |||||
|
MBD5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.020878 (rank : 110) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
SFRP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.014486 (rank : 133) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N474, O00546, O14779 | Gene names | SFRP1, FRP, FRP1, SARP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 1 precursor (sFRP-1) (Frizzled- related protein 1) (FRP-1) (Secreted apoptosis-related protein 2) (SARP-2). | |||||
|
TBD_HUMAN
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.008308 (rank : 157) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UJT1, Q9BWG9, Q9H7Z8 | Gene names | TUBD1, TUBD | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin delta chain (Delta tubulin). | |||||
|
TS101_MOUSE
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.031009 (rank : 94) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61187 | Gene names | Tsg101 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
BCL7C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.013572 (rank : 137) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08664, Q99LS2 | Gene names | Bcl7c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C (B-cell chronic lymphocytic leukemia/lymphoma 7C protein). | |||||
|
CCM2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.018957 (rank : 114) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2Y9 | Gene names | Ccm2, Osm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Malcavernin (Cerebral cavernous malformations protein 2 homolog) (Osmosensing scaffold for MEKK3). | |||||
|
CEP63_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.012114 (rank : 143) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
CN037_MOUSE
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.014961 (rank : 129) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
CT026_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.019332 (rank : 112) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NHU2, Q5TE18, Q8N5R9, Q96M59, Q9BQL2, Q9H127, Q9H128, Q9NQH4, Q9UFV8, Q9Y4V7 | Gene names | C20orf26 | |||
|
Domain Architecture |
|
|||||
| Description | Uncharacterized protein C20orf26. | |||||
|
GAS8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.010888 (rank : 149) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
MLH1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.012835 (rank : 141) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JK91, Q62454 | Gene names | Mlh1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh1 (MutL protein homolog 1). | |||||
|
PAR14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.009421 (rank : 152) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q2EMV9, Q571C6, Q8BN35, Q8VDT6 | Gene names | Parp14, Kiaa1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (Collaborator of STAT6) (CoaSt6). | |||||
|
PO6F1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.002710 (rank : 171) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
PTRF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.016049 (rank : 125) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O54724 | Gene names | Ptrf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polymerase I and transcript release factor. | |||||
|
RABE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.015642 (rank : 127) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
SFRP5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.014161 (rank : 134) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WU66, Q8K269 | Gene names | Sfrp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 5 precursor (sFRP-5). | |||||
|
ANR15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.007646 (rank : 158) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14678, Q5W0W0, Q8IY65, Q8WX74 | Gene names | ANKRD15, KANK, KIAA0172 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 15 (Kidney ankyrin repeat- containing protein). | |||||
|
BRCA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.006693 (rank : 160) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38398 | Gene names | BRCA1, RNF53 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein (RING finger protein 53). | |||||
|
BRD3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.005543 (rank : 167) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15059, Q5T1R7, Q8N5M3, Q92645 | Gene names | BRD3, KIAA0043, RING3L | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (RING3-like protein). | |||||
|
CAC1I_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.003399 (rank : 170) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CCM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.017187 (rank : 120) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BSQ5, Q71RE5, Q8TAT4 | Gene names | CCM2, C7orf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Malcavernin (Cerebral cavernous malformations 2 protein). | |||||
|
DNAI2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.009060 (rank : 154) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9GZS0, Q9H179, Q9NT53 | Gene names | DNAI2 | |||
|
Domain Architecture |
|
|||||
| Description | Dynein intermediate chain 2, axonemal (Axonemal dynein intermediate chain 2). | |||||
|
EMR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.000743 (rank : 174) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14246, Q8NGA7 | Gene names | EMR1, TM7LN3 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 1 precursor (Cell surface glycoprotein EMR1) (EMR1 hormone receptor). | |||||
|
EMR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.000772 (rank : 173) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61549 | Gene names | Emr1, Gpf480 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 1 precursor (Cell surface glycoprotein EMR1) (EMR1 hormone receptor) (Cell surface glycoprotein F4/80). | |||||
|
IF122_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.009114 (rank : 153) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HBG6 | Gene names | IFT122, WDR10, WDR140 | |||
|
Domain Architecture |
|
|||||
| Description | Intraflagellar transport 122 homolog (WD repeat protein 10). | |||||
|
K0859_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.012169 (rank : 142) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N6R0, O94940, Q5TGP9, Q5TGQ0, Q8N2P8, Q96J11, Q96SQ0, Q9Y2Z1, Q9Y3M6 | Gene names | KIAA0859 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0859. | |||||
|
K1C14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.004553 (rank : 169) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P02533, Q14715, Q53XY3, Q9BUE3, Q9UBN2, Q9UBN3, Q9UCY4 | Gene names | KRT14 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cytoskeletal 14 (Cytokeratin-14) (CK-14) (Keratin-14) (K14). | |||||
|
MBN3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.009759 (rank : 150) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUK0, Q5JXN8, Q5JXN9, Q5JXP4, Q6UDQ1, Q8TAD9, Q8TAF4, Q9H0Z7, Q9UF37 | Gene names | MBNL3, CHCR, MBLX39, MBXL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein HCHCR). | |||||
|
MPDZ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.002587 (rank : 172) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75970, O43798, Q5CZ80, Q5JTX3, Q5JTX6, Q5JTX7, Q5JUC3, Q5JUC4, Q5VZ62, Q8N790 | Gene names | MPDZ, MUPP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ- domain protein 1). | |||||
|
MTMR9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.005846 (rank : 164) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2D0, Q80XL4 | Gene names | Mtmr9, Mtmr3 | |||
|
Domain Architecture |
|
|||||
| Description | Myotubularin-related protein 9. | |||||
|
O5BF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | -0.004585 (rank : 177) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NHC7 | Gene names | OR5BF1 | |||
|
Domain Architecture |
|
|||||
| Description | Olfactory receptor 5BF1. | |||||
|
P5CS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.021351 (rank : 108) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54886, O95952, Q9UM72 | Gene names | ALDH18A1, P5CS, PYCS | |||
|
Domain Architecture |

|
|||||
| Description | Delta 1-pyrroline-5-carboxylate synthetase (P5CS) (Aldehyde dehydrogenase 18 family member A1) [Includes: Glutamate 5-kinase (EC 2.7.2.11) (Gamma-glutamyl kinase) (GK); Gamma-glutamyl phosphate reductase (GPR) (EC 1.2.1.41) (Glutamate-5-semialdehyde dehydrogenase) (Glutamyl-gamma-semialdehyde dehydrogenase)]. | |||||
|
RPGF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.005621 (rank : 165) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4G8 | Gene names | RAPGEF2, KIAA0313, NRAPGEP, PDZGEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rap guanine nucleotide exchange factor 2 (Neural RAP guanine nucleotide exchange protein) (nRap GEP) (PDZ domain-containing guanine nucleotide exchange factor 1) (PDZ-GEF1) (RA-GEF). | |||||
|
STX1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.008460 (rank : 156) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16623, O15447, O15448, Q12936, Q9BPZ6 | Gene names | STX1A, STX1 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-1A (Neuron-specific antigen HPC-1). | |||||
|
STX1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.008618 (rank : 155) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35526 | Gene names | Stx1a | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-1A (Neuron-specific antigen HPC-1). | |||||
|
TACC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.011638 (rank : 145) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
CAR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.146736 (rank : 73) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NPP4, Q96J81, Q96J82, Q96J83 | Gene names | CARD12, CLAN, CLAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 12 (Ice protease- activating factor) (Ipaf) (CARD, LRR, and NACHT-containing protein) (Clan protein). | |||||
|
CGRF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.058260 (rank : 81) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99675, Q96BX2 | Gene names | CGRRF1, CGR19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with RING finger domain 1 (Cell growth regulatory gene 19 protein). | |||||
|
CGRF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.056264 (rank : 82) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BMJ7, Q3U3F1, Q8CI24 | Gene names | Cgrrf1, Cgr19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with RING finger domain 1 (Cell growth regulatory gene 19 protein). | |||||
|
FTS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.159868 (rank : 72) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H8T0 | Gene names | FTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fused toes protein homolog (Ft1). | |||||
|
FTS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.162635 (rank : 71) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q64362 | Gene names | Fts, Fif, Ft1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fused toes protein (FT1). | |||||
|
MYLIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.061224 (rank : 80) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WY64, Q9BU73, Q9NRL9, Q9UHE7 | Gene names | MYLIP, BZF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
RFFL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.084372 (rank : 78) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WZ73, Q8NHW0, Q8TBY7, Q96BE6 | Gene names | RFFL, RNF189, RNF34L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (FYVE-RING finger protein Sakura) (Fring) (Caspases-8 and -10- associated RING finger protein 2) (CARP-2) (Caspase regulator CARP2) (RING finger protein 189) (RING finger protein 34-like). | |||||
|
RFFL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.085913 (rank : 75) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZQM0, Q5SVC2, Q5SVC4, Q9D543, Q9D9B1 | Gene names | Rffl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (Fring). | |||||
|
RKHD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.063328 (rank : 79) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86XN8 | Gene names | RKHD1, KIAA2031 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and KH domain-containing protein 1. | |||||
|
RNF26_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.094853 (rank : 74) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BY78 | Gene names | RNF26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 26. | |||||
|
RNF34_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.084419 (rank : 77) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q969K3, Q8NG47, Q9H6W8 | Gene names | RNF34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 34 (RING finger protein RIFF) (FYVE-RING finger protein Momo) (Human RING finger homologous to inhibitor of apoptosis protein) (hRFI) (Caspases-8 and -10-associated RING finger protein 1) (CARP-1) (Caspase regulator CARP1). | |||||
|
RNF34_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.085521 (rank : 76) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99KR6, Q3UV45 | Gene names | Rnf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 34 (RING finger protein RIFF) (Phafin-1). | |||||
|
UB2V1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.245509 (rank : 69) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13404, Q13403, Q13532, Q96H34, Q9GZT0, Q9GZW1, Q9H4J3, Q9H4J4, Q9UKL1, Q9UM48, Q9UM49, Q9UM50 | Gene names | UBE2V1, CROC1, UBE2V, UEV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 variant 1 (UEV-1) (CROC-1) (Ubiquitin- conjugating enzyme variant Kua) (TRAF6-regulated IKK activator 1 beta Uev1A). | |||||
|
UB2V1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.242782 (rank : 70) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CZY3, Q8VEB5, Q9ERI7 | Gene names | Ube2v1, Croc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 variant 1 (UEV-1) (CROC-1). | |||||
|
UB2V2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.251330 (rank : 68) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15819 | Gene names | UBE2V2, MMS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 variant 2 (MMS2) (Enterocyte differentiation-associated factor EDAF-1) (Enterocyte differentiation- promoting factor) (EDPF-1) (Vitamin D3-inducible protein) (DDVit 1). | |||||
|
UB2V2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.254792 (rank : 67) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9D2M8, Q8BGH6, Q8CE99, Q8K2V7, Q9CYD7, Q9ERI8 | Gene names | Ube2v2, Mms2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 variant 2 (Ubc-like protein MMS2). | |||||
|
BIRC6_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 161 | |
| SwissProt Accessions | Q9NR09, Q9ULD1 | Gene names | BIRC6, KIAA1289 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 6 (Ubiquitin-conjugating BIR-domain enzyme apollon). | |||||
|
UB2G2_HUMAN
|
||||||
| NC score | 0.608468 (rank : 2) | θ value | 1.25267e-09 (rank : 13) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P60604, P56554 | Gene names | UBE2G2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 G2 (EC 6.3.2.19) (Ubiquitin-protein ligase G2) (Ubiquitin carrier protein G2). | |||||
|
UB2G2_MOUSE
|
||||||
| NC score | 0.608468 (rank : 3) | θ value | 1.25267e-09 (rank : 14) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P60605, P56554 | Gene names | Ube2g2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 G2 (EC 6.3.2.19) (Ubiquitin-protein ligase G2) (Ubiquitin carrier protein G2). | |||||
|
UBE2A_HUMAN
|
||||||
| NC score | 0.606817 (rank : 4) | θ value | 4.59992e-12 (rank : 4) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P49459, Q4TTG1, Q96FX4 | Gene names | UBE2A, RAD6A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 A (EC 6.3.2.19) (Ubiquitin-protein ligase A) (Ubiquitin carrier protein A) (HR6A) (hHR6A). | |||||
|
UBE2A_MOUSE
|
||||||
| NC score | 0.606817 (rank : 5) | θ value | 4.59992e-12 (rank : 5) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Z255 | Gene names | Ube2a, Rad6a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 A (EC 6.3.2.19) (Ubiquitin-protein ligase A) (Ubiquitin carrier protein A) (HR6A) (mHR6A). | |||||
|
UBE2B_HUMAN
|
||||||
| NC score | 0.605314 (rank : 6) | θ value | 1.74796e-11 (rank : 7) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P63146, P23567, Q4PJ15, Q9D0J6 | Gene names | UBE2B, RAD6B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 B (EC 6.3.2.19) (Ubiquitin-protein ligase B) (Ubiquitin carrier protein B) (HR6B) (hHR6B) (E2-17 kDa). | |||||
|
UBE2B_MOUSE
|
||||||
| NC score | 0.605314 (rank : 7) | θ value | 1.74796e-11 (rank : 8) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P63147, P23567, Q3UGS2, Q9D0J6 | Gene names | Ube2b, Rad6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 B (EC 6.3.2.19) (Ubiquitin-protein ligase B) (Ubiquitin carrier protein B) (HR6B) (E214K). | |||||
|
UBC9_HUMAN
|
||||||
| NC score | 0.601797 (rank : 8) | θ value | 4.76016e-09 (rank : 22) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P63279, P50550, Q15698, Q86VB3 | Gene names | UBE2I, UBC9, UBCE9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SUMO-1-conjugating enzyme UBC9 (EC 6.3.2.19) (SUMO-1-protein ligase) (Ubiquitin-conjugating enzyme E2 I) (Ubiquitin-protein ligase I) (Ubiquitin carrier protein I) (Ubiquitin carrier protein 9) (p18). | |||||
|
UBC9_MOUSE
|
||||||
| NC score | 0.601797 (rank : 9) | θ value | 4.76016e-09 (rank : 23) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P63280, P50550, Q15698, Q86VB3 | Gene names | Ube2i, Ubc9, Ubce9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SUMO-1-conjugating enzyme UBC9 (EC 6.3.2.19) (SUMO-1-protein ligase) (Ubiquitin-conjugating enzyme E2 I) (Ubiquitin-protein ligase I) (Ubiquitin carrier protein I) (Ubiquitin carrier protein 9) (mUBC9). | |||||
|
UB2R1_MOUSE
|
||||||
| NC score | 0.598701 (rank : 10) | θ value | 2.13673e-09 (rank : 17) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8CFI2, Q505K8 | Gene names | Cdc34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2-32 kDa complementing (EC 6.3.2.19) (Ubiquitin-protein ligase) (Ubiquitin carrier protein) (E2-CDC34). | |||||
|
UB2R1_HUMAN
|
||||||
| NC score | 0.598606 (rank : 11) | θ value | 2.13673e-09 (rank : 16) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P49427 | Gene names | CDC34, UBE2R1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2-32 kDa complementing (EC 6.3.2.19) (Ubiquitin-protein ligase) (Ubiquitin carrier protein) (E2-CDC34). | |||||
|
UB2G1_HUMAN
|
||||||
| NC score | 0.597347 (rank : 12) | θ value | 2.79066e-09 (rank : 18) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P62253, Q99462 | Gene names | UBE2G1, UBE2G | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 G1 (EC 6.3.2.19) (Ubiquitin-protein ligase G1) (Ubiquitin carrier protein G1) (E217K) (UBC7). | |||||
|
UB2G1_MOUSE
|
||||||
| NC score | 0.597347 (rank : 13) | θ value | 2.79066e-09 (rank : 19) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P62254, Q99462 | Gene names | Ube2g1, Ube2g | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 G1 (EC 6.3.2.19) (Ubiquitin-protein ligase G1) (Ubiquitin carrier protein G1) (E217K) (UBC7). | |||||
|
UBE2T_HUMAN
|
||||||
| NC score | 0.596853 (rank : 14) | θ value | 1.47974e-10 (rank : 10) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NPD8 | Gene names | UBE2T | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 T (EC 6.3.2.19) (Ubiquitin-protein ligase T) (Ubiquitin carrier protein T). | |||||
|
UBE2N_HUMAN
|
||||||
| NC score | 0.594026 (rank : 15) | θ value | 6.21693e-09 (rank : 24) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P61088, Q16781 | Gene names | UBE2N, BLU | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 N (EC 6.3.2.19) (Ubiquitin-protein ligase N) (Ubiquitin carrier protein N) (Ubc13) (Bendless-like ubiquitin-conjugating enzyme). | |||||
|
UBE2N_MOUSE
|
||||||
| NC score | 0.594026 (rank : 16) | θ value | 6.21693e-09 (rank : 25) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P61089, Q16781, Q6ZWZ0, Q9DAJ6 | Gene names | Ube2n, Blu | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 N (EC 6.3.2.19) (Ubiquitin-protein ligase N) (Ubiquitin carrier protein N) (Ubc13) (Bendless-like ubiquitin-conjugating enzyme). | |||||
|
UBE2T_MOUSE
|
||||||
| NC score | 0.590668 (rank : 17) | θ value | 3.64472e-09 (rank : 20) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9CQ37 | Gene names | Ube2t | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 T (EC 6.3.2.19) (Ubiquitin-protein ligase T) (Ubiquitin carrier protein T). | |||||
|
UBE2S_MOUSE
|
||||||
| NC score | 0.588078 (rank : 18) | θ value | 1.53129e-07 (rank : 34) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q921J4, Q3U4V7 | Gene names | Ube2s, E2epf | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 S (EC 6.3.2.19) (Ubiquitin-protein ligase S) (Ubiquitin carrier protein S) (Ubiquitin-conjugating enzyme E2-24 kDa) (E2-EPF5). | |||||
|
UBE2S_HUMAN
|
||||||
| NC score | 0.587950 (rank : 19) | θ value | 1.53129e-07 (rank : 33) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16763, Q9BTC1 | Gene names | UBE2S, E2EPF | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 S (EC 6.3.2.19) (Ubiquitin-protein ligase S) (Ubiquitin carrier protein S) (Ubiquitin-conjugating enzyme E2-24 kDa) (E2-EPF5). | |||||
|
UB2L6_MOUSE
|
||||||
| NC score | 0.585967 (rank : 20) | θ value | 6.87365e-08 (rank : 31) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9QZU9, Q922F1, Q9CQN0 | Gene names | Ube2l6, Ubce8 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 L6 (EC 6.3.2.19) (Ubiquitin-protein ligase L6) (Ubiquitin carrier protein L6) (UbcM8). | |||||
|
UB2D2_HUMAN
|
||||||
| NC score | 0.575811 (rank : 21) | θ value | 4.0297e-08 (rank : 27) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P62837, P51669 | Gene names | UBE2D2, UBC4, UBCH5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 D2 (EC 6.3.2.19) (Ubiquitin-protein ligase D2) (Ubiquitin carrier protein D2) (Ubiquitin-conjugating enzyme E2-17 kDa 2) (E2(17)KB 2). | |||||
|
UB2D2_MOUSE
|
||||||
| NC score | 0.575811 (rank : 22) | θ value | 4.0297e-08 (rank : 28) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P62838, P51669 | Gene names | Ube2d2, Ubc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 D2 (EC 6.3.2.19) (Ubiquitin-protein ligase D2) (Ubiquitin carrier protein D2) (Ubiquitin-conjugating enzyme E2-17 kDa 2) (E2(17)KB 2). | |||||
|
UB2D3_HUMAN
|
||||||
| NC score | 0.575688 (rank : 23) | θ value | 4.0297e-08 (rank : 29) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P61077, P47986, Q6IB88, Q6NXS4 | Gene names | UBE2D3, UBCH5C | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 D3 (EC 6.3.2.19) (Ubiquitin-protein ligase D3) (Ubiquitin carrier protein D3) (Ubiquitin-conjugating enzyme E2-17 kDa 3) (E2(17)KB 3). | |||||
|
UB2D3_MOUSE
|
||||||
| NC score | 0.575688 (rank : 24) | θ value | 4.0297e-08 (rank : 30) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P61079 | Gene names | Ube2d3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 D3 (EC 6.3.2.19) (Ubiquitin-protein ligase D3) (Ubiquitin carrier protein D3) (Ubiquitin-conjugating enzyme E2-17 kDa 3) (E2(17)KB 3). | |||||
|
UB2L3_HUMAN
|
||||||
| NC score | 0.575233 (rank : 25) | θ value | 6.43352e-06 (rank : 43) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P68036, P51966, P70653, Q9HAV1 | Gene names | UBE2L3, UBCE7, UBCH7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 L3 (EC 6.3.2.19) (Ubiquitin-protein ligase L3) (Ubiquitin carrier protein L3) (UbcH7) (E2-F1) (L-UBC). | |||||
|
UB2L3_MOUSE
|
||||||
| NC score | 0.575233 (rank : 26) | θ value | 6.43352e-06 (rank : 44) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P68037, P51966, P70653, Q9HAV1 | Gene names | Ube2l3, Ubce7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 L3 (EC 6.3.2.19) (Ubiquitin-protein ligase L3) (Ubiquitin carrier protein L3) (UbcM4). | |||||
|
UB2D1_HUMAN
|
||||||
| NC score | 0.572369 (rank : 27) | θ value | 3.41135e-07 (rank : 35) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P51668 | Gene names | UBE2D1, UBCH5, UBCH5A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 D1 (EC 6.3.2.19) (Ubiquitin-protein ligase D1) (Ubiquitin carrier protein D1) (UbcH5) (Ubiquitin- conjugating enzyme E2-17 kDa 1) (E2(17)KB 1). | |||||
|
UB2D1_MOUSE
|
||||||
| NC score | 0.572369 (rank : 28) | θ value | 3.41135e-07 (rank : 36) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P61080 | Gene names | Ube2d1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 D1 (EC 6.3.2.19) (Ubiquitin-protein ligase D1) (Ubiquitin carrier protein D1) (Ubiquitin-conjugating enzyme E2-17 kDa 1) (E2(17)KB 1). | |||||
|
UB2L6_HUMAN
|
||||||
| NC score | 0.569208 (rank : 29) | θ value | 2.44474e-05 (rank : 46) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O14933, Q9UEZ0 | Gene names | UBE2L6, UBCH8 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 L6 (EC 6.3.2.19) (Ubiquitin-protein ligase L6) (Ubiquitin carrier protein L6) (UbcH8) (Retinoic acid- induced gene B protein) (RIG-B). | |||||
|
UBE2C_HUMAN
|
||||||
| NC score | 0.568887 (rank : 30) | θ value | 1.43324e-05 (rank : 45) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O00762 | Gene names | UBE2C, UBCH10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 C (EC 6.3.2.19) (Ubiquitin-protein ligase C) (Ubiquitin carrier protein C) (UbcH10). | |||||
|
UBE2C_MOUSE
|
||||||
| NC score | 0.564860 (rank : 31) | θ value | 9.29e-05 (rank : 53) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9D1C1, Q3TUJ0 | Gene names | Ube2c, Ubch10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 C (EC 6.3.2.19) (Ubiquitin-protein ligase C) (Ubiquitin carrier protein C) (UbcH10). | |||||
|
UBC1_HUMAN
|
||||||
| NC score | 0.554990 (rank : 32) | θ value | 5.44631e-05 (rank : 51) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P61086, O54806, P27924, Q16721, Q9CVV9, Q9Y2D3 | Gene names | HIP2, LIG | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2-25 kDa (EC 6.3.2.19) (Ubiquitin- protein ligase) (Ubiquitin carrier protein) (E2(25K)) (Huntingtin- interacting protein 2) (HIP-2). | |||||
|
UBC1_MOUSE
|
||||||
| NC score | 0.554990 (rank : 33) | θ value | 5.44631e-05 (rank : 52) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P61087, O54806, P27924, Q16721, Q9CVV9 | Gene names | Hip2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2-25 kDa (EC 6.3.2.19) (Ubiquitin- protein ligase) (Ubiquitin carrier protein) (E2(25K)) (Huntingtin- interacting protein 2) (HIP-2). | |||||
|
UB2E3_HUMAN
|
||||||
| NC score | 0.551164 (rank : 34) | θ value | 3.19293e-05 (rank : 47) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q969T4, Q5U0R7, Q7Z4W4 | Gene names | UBE2E3, UBCE4, UBCH9 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 E3 (EC 6.3.2.19) (Ubiquitin-protein ligase E3) (Ubiquitin carrier protein E3) (Ubiquitin-conjugating enzyme E2-23 kDa) (UbcH9) (UbcM2). | |||||
|
UB2E3_MOUSE
|
||||||
| NC score | 0.551164 (rank : 35) | θ value | 3.19293e-05 (rank : 48) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P52483, O09180, Q3TI00, Q91X63 | Gene names | Ube2e3, Ubce4, Ubcm2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 E3 (EC 6.3.2.19) (Ubiquitin-protein ligase E3) (Ubiquitin carrier protein E3) (Ubiquitin-conjugating enzyme E2-23 kDa) (UbcM2). | |||||
|
UB2E2_HUMAN
|
||||||
| NC score | 0.549768 (rank : 36) | θ value | 4.1701e-05 (rank : 49) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96LR5 | Gene names | UBE2E2, UBCH8 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 E2 (EC 6.3.2.19) (Ubiquitin-protein ligase E2) (Ubiquitin carrier protein E2) (UbcH8). | |||||
|
UB2E2_MOUSE
|
||||||
| NC score | 0.548978 (rank : 37) | θ value | 4.1701e-05 (rank : 50) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q91W82 | Gene names | Ube2e2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 E2 (EC 6.3.2.19) (Ubiquitin-protein ligase E2) (Ubiquitin carrier protein E2). | |||||
|
UB2E1_HUMAN
|
||||||
| NC score | 0.547314 (rank : 38) | θ value | 0.000121331 (rank : 54) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P51965 | Gene names | UBE2E1, UBCH6 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 E1 (EC 6.3.2.19) (Ubiquitin-protein ligase E1) (Ubiquitin carrier protein E1) (UbcH6). | |||||
|
UB2E1_MOUSE
|
||||||
| NC score | 0.545927 (rank : 39) | θ value | 0.000121331 (rank : 55) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P52482 | Gene names | Ube2e1, Ubce5, Ubcm3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 E1 (EC 6.3.2.19) (Ubiquitin-protein ligase E1) (Ubiquitin carrier protein E1) (UbcM3). | |||||
|
UBC12_HUMAN
|
||||||
| NC score | 0.542311 (rank : 40) | θ value | 0.000786445 (rank : 56) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P61081, O76069, Q8VC50 | Gene names | UBE2M, UBC12 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD8-conjugating enzyme Ubc12 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme E2 M) (NEDD8 protein ligase) (NEDD8 carrier protein). | |||||
|
UBC12_MOUSE
|
||||||
| NC score | 0.542311 (rank : 41) | θ value | 0.000786445 (rank : 57) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P61082, O76069, Q8VC50 | Gene names | Ube2m, Ubc-rs2, Ubc12 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD8-conjugating enzyme Ubc12 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme E2 M) (NEDD8 protein ligase) (NEDD8 carrier protein). | |||||
|
UBE2H_HUMAN
|
||||||
| NC score | 0.533438 (rank : 42) | θ value | 0.0563607 (rank : 63) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P62256, P37286 | Gene names | UBE2H | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 H (EC 6.3.2.19) (Ubiquitin-protein ligase H) (Ubiquitin carrier protein H) (UbcH2) (E2-20K). | |||||
|
UBE2H_MOUSE
|
||||||
| NC score | 0.533438 (rank : 43) | θ value | 0.0563607 (rank : 64) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P62257, P37286 | Gene names | Ube2h | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 H (EC 6.3.2.19) (Ubiquitin-protein ligase H) (Ubiquitin carrier protein H) (UBCH2) (E2-20K). | |||||
|
UBE2U_HUMAN
|
||||||
| NC score | 0.515188 (rank : 44) | θ value | 0.62314 (rank : 77) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5VVX9, Q8N1D4 | Gene names | UBE2U | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 U (EC 6.3.2.19) (Ubiquitin-protein ligase U) (Ubiquitin carrier protein U). | |||||
|
UBE2W_MOUSE
|
||||||
| NC score | 0.511142 (rank : 45) | θ value | 0.0736092 (rank : 65) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8VDW4, Q9D5H3 | Gene names | Ube2w | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin-conjugating enzyme E2 W (EC 6.3.2.19) (Ubiquitin- protein ligase W) (Ubiquitin carrier protein W). | |||||
|
UBE2W_HUMAN
|
||||||
| NC score | 0.509921 (rank : 46) | θ value | 0.0961366 (rank : 66) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96B02, Q9H823, Q9HAG6, Q9NV07 | Gene names | UBE2W | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin-conjugating enzyme E2 W (EC 6.3.2.19) (Ubiquitin- protein ligase W) (Ubiquitin carrier protein W). | |||||
|
UB2J2_MOUSE
|
||||||
| NC score | 0.488205 (rank : 47) | θ value | 0.00298849 (rank : 59) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6P073, Q8C6A1, Q91Y64, Q9CWY5, Q9DC18, Q9QX58 | Gene names | Ube2j2, Ncube2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 J2 (EC 6.3.2.19) (Non-canonical ubiquitin-conjugating enzyme 2) (NCUBE2). | |||||
|
UB2J2_HUMAN
|
||||||
| NC score | 0.486000 (rank : 48) | θ value | 0.00298849 (rank : 58) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8N2K1, Q96N26, Q96T84 | Gene names | UBE2J2, NCUBE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 J2 (EC 6.3.2.19) (Non-canonical ubiquitin-conjugating enzyme 2) (NCUBE2). | |||||
|
BIRC5_MOUSE
|
||||||
| NC score | 0.478743 (rank : 49) | θ value | 1.33837e-11 (rank : 6) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70201, Q923F7, Q9WU53, Q9WU54 | Gene names | Birc5, Api4, Iap4 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 5 (Apoptosis inhibitor survivin) (Apoptosis inhibitor 4) (TIAP). | |||||
|
BIRC5_HUMAN
|
||||||
| NC score | 0.473040 (rank : 50) | θ value | 8.67504e-11 (rank : 9) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15392, Q2I3N8, Q4VGX0, Q53F61, Q5MGC6, Q6FHL2, Q75SP2, Q9P2W8 | Gene names | BIRC5, API4, IAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 5 (Apoptosis inhibitor survivin) (Apoptosis inhibitor 4). | |||||
|
UB2J1_HUMAN
|
||||||
| NC score | 0.451044 (rank : 51) | θ value | 0.0113563 (rank : 60) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y385, Q5W0N4, Q9BZ32, Q9NQL3, Q9NY66, Q9P011, Q9P0S0, Q9UF10 | Gene names | UBE2J1, NCUBE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 J1 (EC 6.3.2.19) (Non-canonical ubiquitin-conjugating enzyme 1) (NCUBE1) (Yeast ubiquitin-conjugating enzyme UBC6 homolog E) (HSUBC6e). | |||||
|
UB2J1_MOUSE
|
||||||
| NC score | 0.450171 (rank : 52) | θ value | 0.0113563 (rank : 61) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JJZ4, Q9DC92 | Gene names | Ube2j1, Ncube1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 J1 (EC 6.3.2.19) (Non-canonical ubiquitin-conjugating enzyme 1) (NCUBE1). | |||||
|
BIRC4_HUMAN
|
||||||
| NC score | 0.410027 (rank : 53) | θ value | 2.0648e-12 (rank : 2) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P98170, Q9NQ14 | Gene names | BIRC4, API3, IAP3, XIAP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP-like protein) (HILP). | |||||
|
BIRC4_MOUSE
|
||||||
| NC score | 0.409424 (rank : 54) | θ value | 4.59992e-12 (rank : 3) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60989, O08865 | Gene names | Birc4, Aipa, Api3, Miha, Xiap | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP homolog A) (MIAP3) (MIAP-3). | |||||
|
BIRC3_HUMAN
|
||||||
| NC score | 0.380896 (rank : 55) | θ value | 9.59137e-10 (rank : 12) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13489, Q16628, Q9HC27, Q9UP46 | Gene names | BIRC3, API2, IAP1, MIHC | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (HIAP1) (HIAP-1) (C-IAP2) (TNFR2-TRAF signaling complex protein 1) (IAP homolog C) (Apoptosis inhibitor 2) (API2). | |||||
|
BIRC2_HUMAN
|
||||||
| NC score | 0.380637 (rank : 56) | θ value | 9.59137e-10 (rank : 11) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13490, Q16516, Q4TTG0 | Gene names | BIRC2, API1, IAP2, MIHB | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (HIAP2) (HIAP-2) (C-IAP1) (TNFR2-TRAF signaling complex protein 2) (IAP homolog B). | |||||
|
BIRC2_MOUSE
|
||||||
| NC score | 0.379131 (rank : 57) | θ value | 1.63604e-09 (rank : 15) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62210, O08864 | Gene names | Birc2, Birc3, Iap2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (MIAP2) (MIAP-2). | |||||
|
BIRC3_MOUSE
|
||||||
| NC score | 0.378831 (rank : 58) | θ value | 4.76016e-09 (rank : 21) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08863 | Gene names | Birc3, Birc2, Iap1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (MIAP1) (MIAP-1). | |||||
|
BIRC8_HUMAN
|
||||||
| NC score | 0.371664 (rank : 59) | θ value | 6.43352e-06 (rank : 42) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96P09, Q96RW5 | Gene names | BIRC8, ILP2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 8 (Inhibitor of apoptosis- like protein 2) (IAP-like protein 2) (ILP-2) (Testis-specific inhibitor of apoptosis). | |||||
|
BIRC7_HUMAN
|
||||||
| NC score | 0.354464 (rank : 60) | θ value | 1.53129e-07 (rank : 32) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96CA5, Q9BQV0, Q9H2A8, Q9HAP7 | Gene names | BIRC7, KIAP, LIVIN, MLIAP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 7 (Kidney inhibitor of apoptosis protein) (KIAP) (Melanoma inhibitor of apoptosis protein) (ML-IAP) (Livin). | |||||
|
BIR1B_MOUSE
|
||||||
| NC score | 0.332930 (rank : 61) | θ value | 4.0297e-08 (rank : 26) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QUK4, O09124, Q9R030 | Gene names | Birc1b, Naip-rs6, Naip2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1b (Neuronal apoptosis inhibitory protein 2). | |||||
|
BIRC1_HUMAN
|
||||||
| NC score | 0.331518 (rank : 62) | θ value | 7.59969e-07 (rank : 37) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13075, O75857, Q13730, Q99796 | Gene names | BIRC1, NAIP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1 (Neuronal apoptosis inhibitory protein). | |||||
|
BIR1E_MOUSE
|
||||||
| NC score | 0.325691 (rank : 63) | θ value | 2.88788e-06 (rank : 40) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9R016, O09121, O09122, P81703, Q9R029 | Gene names | Birc1e, Naip-rs3, Naip5 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1e (Neuronal apoptosis inhibitory protein 5). | |||||
|
BIR1F_MOUSE
|
||||||
| NC score | 0.325611 (rank : 64) | θ value | 2.21117e-06 (rank : 38) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JIB6, O09121, O09122, P81704 | Gene names | Birc1f, Naip-rs4, Naip6 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1f (Neuronal apoptosis inhibitory protein 6). | |||||
|
BIR1G_MOUSE
|
||||||
| NC score | 0.325202 (rank : 65) | θ value | 2.88788e-06 (rank : 41) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JIB3 | Gene names | Birc1g, Naip7 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1g (Neuronal apoptosis inhibitory protein 7). | |||||
|
BIR1A_MOUSE
|
||||||
| NC score | 0.324270 (rank : 66) | θ value | 2.88788e-06 (rank : 39) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QWK5, Q9JIB5, Q9R017 | Gene names | Birc1a, Naip, Naip1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1a (Neuronal apoptosis inhibitory protein 1). | |||||
|
UB2V2_MOUSE
|
||||||
| NC score | 0.254792 (rank : 67) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9D2M8, Q8BGH6, Q8CE99, Q8K2V7, Q9CYD7, Q9ERI8 | Gene names | Ube2v2, Mms2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 variant 2 (Ubc-like protein MMS2). | |||||
|
UB2V2_HUMAN
|
||||||
| NC score | 0.251330 (rank : 68) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15819 | Gene names | UBE2V2, MMS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 variant 2 (MMS2) (Enterocyte differentiation-associated factor EDAF-1) (Enterocyte differentiation- promoting factor) (EDPF-1) (Vitamin D3-inducible protein) (DDVit 1). | |||||
|
UB2V1_HUMAN
|
||||||
| NC score | 0.245509 (rank : 69) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13404, Q13403, Q13532, Q96H34, Q9GZT0, Q9GZW1, Q9H4J3, Q9H4J4, Q9UKL1, Q9UM48, Q9UM49, Q9UM50 | Gene names | UBE2V1, CROC1, UBE2V, UEV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 variant 1 (UEV-1) (CROC-1) (Ubiquitin- conjugating enzyme variant Kua) (TRAF6-regulated IKK activator 1 beta Uev1A). | |||||
|
UB2V1_MOUSE
|
||||||
| NC score | 0.242782 (rank : 70) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CZY3, Q8VEB5, Q9ERI7 | Gene names | Ube2v1, Croc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 variant 1 (UEV-1) (CROC-1). | |||||
|
FTS1_MOUSE
|
||||||
| NC score | 0.162635 (rank : 71) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q64362 | Gene names | Fts, Fif, Ft1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fused toes protein (FT1). | |||||
|
FTS1_HUMAN
|
||||||
| NC score | 0.159868 (rank : 72) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H8T0 | Gene names | FTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fused toes protein homolog (Ft1). | |||||
|
CAR12_HUMAN
|
||||||
| NC score | 0.146736 (rank : 73) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NPP4, Q96J81, Q96J82, Q96J83 | Gene names | CARD12, CLAN, CLAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 12 (Ice protease- activating factor) (Ipaf) (CARD, LRR, and NACHT-containing protein) (Clan protein). | |||||
|
RNF26_HUMAN
|
||||||
| NC score | 0.094853 (rank : 74) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BY78 | Gene names | RNF26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 26. | |||||
|
RFFL_MOUSE
|
||||||
| NC score | 0.085913 (rank : 75) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZQM0, Q5SVC2, Q5SVC4, Q9D543, Q9D9B1 | Gene names | Rffl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (Fring). | |||||
|
RNF34_MOUSE
|
||||||
| NC score | 0.085521 (rank : 76) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99KR6, Q3UV45 | Gene names | Rnf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 34 (RING finger protein RIFF) (Phafin-1). | |||||
|
RNF34_HUMAN
|
||||||
| NC score | 0.084419 (rank : 77) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q969K3, Q8NG47, Q9H6W8 | Gene names | RNF34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 34 (RING finger protein RIFF) (FYVE-RING finger protein Momo) (Human RING finger homologous to inhibitor of apoptosis protein) (hRFI) (Caspases-8 and -10-associated RING finger protein 1) (CARP-1) (Caspase regulator CARP1). | |||||
|
RFFL_HUMAN
|
||||||
| NC score | 0.084372 (rank : 78) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WZ73, Q8NHW0, Q8TBY7, Q96BE6 | Gene names | RFFL, RNF189, RNF34L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rififylin (RING finger and FYVE-like domain-containing protein 1) (FYVE-RING finger protein Sakura) (Fring) (Caspases-8 and -10- associated RING finger protein 2) (CARP-2) (Caspase regulator CARP2) (RING finger protein 189) (RING finger protein 34-like). | |||||
|
RKHD1_HUMAN
|
||||||
| NC score | 0.063328 (rank : 79) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86XN8 | Gene names | RKHD1, KIAA2031 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and KH domain-containing protein 1. | |||||
|
MYLIP_HUMAN
|
||||||
| NC score | 0.061224 (rank : 80) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WY64, Q9BU73, Q9NRL9, Q9UHE7 | Gene names | MYLIP, BZF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
CGRF1_HUMAN
|
||||||
| NC score | 0.058260 (rank : 81) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99675, Q96BX2 | Gene names | CGRRF1, CGR19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with RING finger domain 1 (Cell growth regulatory gene 19 protein). | |||||
|
CGRF1_MOUSE
|
||||||
| NC score | 0.056264 (rank : 82) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BMJ7, Q3U3F1, Q8CI24 | Gene names | Cgrrf1, Cgr19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with RING finger domain 1 (Cell growth regulatory gene 19 protein). | |||||
|
SCFD2_MOUSE
|
||||||
| NC score | 0.039910 (rank : 83) | θ value | 0.62314 (rank : 76) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BTY8, Q3USN5, Q8BFV9, Q8BKU5, Q8BKZ0, Q8BTP6, Q8BTQ9, Q8BXI1, Q8BXS0, Q8BZ58 | Gene names | Scfd2, Stxbp1l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sec1 family domain-containing protein 2 (Syntaxin-binding protein 1- like 1) (Neuronal Sec1). | |||||
|
TS101_HUMAN
|
||||||
| NC score | 0.039858 (rank : 84) | θ value | 1.06291 (rank : 87) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99816, Q9BUM5 | Gene names | TSG101 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
USP9Y_HUMAN
|
||||||
| NC score | 0.039410 (rank : 85) | θ value | 0.0961366 (rank : 67) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
CLN3_HUMAN
|
||||||
| NC score | 0.038358 (rank : 86) | θ value | 1.06291 (rank : 82) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13286, O00668, Q9UP09, Q9UP11, Q9UP12, Q9UP13, Q9UP14 | Gene names | CLN3, BTS | |||
|
Domain Architecture |

|
|||||
| Description | Protein CLN3 (Battenin) (Batten disease protein). | |||||
|
WDR75_HUMAN
|
||||||
| NC score | 0.036436 (rank : 87) | θ value | 0.813845 (rank : 81) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWA0, Q96J10, Q9H8U8, Q9H9U5, Q9H9V8, Q9UIX2 | Gene names | WDR75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 75. | |||||
|
FEZ2_HUMAN
|
||||||
| NC score | 0.035797 (rank : 88) | θ value | 0.62314 (rank : 75) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UHY8, Q76LN0, Q99690 | Gene names | FEZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 2 (Zygin-2) (Zygin II). | |||||
|
ARRS_MOUSE
|
||||||
| NC score | 0.034500 (rank : 89) | θ value | 0.279714 (rank : 68) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20443, Q91W62 | Gene names | Sag | |||
|
Domain Architecture |

|
|||||
| Description | S-arrestin (Retinal S-antigen) (48 kDa protein) (S-AG) (Rod photoreceptor arrestin). | |||||
|
IFT74_HUMAN
|
||||||
| NC score | 0.032802 (rank : 90) | θ value | 0.0431538 (rank : 62) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
LRRF1_HUMAN
|
||||||
| NC score | 0.031924 (rank : 91) | θ value | 0.47712 (rank : 72) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
USP9X_MOUSE
|
||||||
| NC score | 0.031155 (rank : 92) | θ value | 2.36792 (rank : 100) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
USP9X_HUMAN
|
||||||
| NC score | 0.031056 (rank : 93) | θ value | 2.36792 (rank : 99) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
TS101_MOUSE
|
||||||
| NC score | 0.031009 (rank : 94) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61187 | Gene names | Tsg101 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
K0423_HUMAN
|
||||||
| NC score | 0.028832 (rank : 95) | θ value | 1.06291 (rank : 84) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
P5CS_MOUSE
|
||||||
| NC score | 0.027896 (rank : 96) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z110, Q9R1P6 | Gene names | Aldh18a1, P5cs, Pycs | |||
|
Domain Architecture |

|
|||||
| Description | Delta 1-pyrroline-5-carboxylate synthetase (P5CS) (Aldehyde dehydrogenase 18 family member A1) [Includes: Glutamate 5-kinase (EC 2.7.2.11) (Gamma-glutamyl kinase) (GK); Gamma-glutamyl phosphate reductase (GPR) (EC 1.2.1.41) (Glutamate-5-semialdehyde dehydrogenase) (Glutamyl-gamma-semialdehyde dehydrogenase)]. | |||||
|
CLSPN_MOUSE
|
||||||
| NC score | 0.026876 (rank : 97) | θ value | 1.38821 (rank : 89) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
IFT74_MOUSE
|
||||||
| NC score | 0.025839 (rank : 98) | θ value | 2.36792 (rank : 96) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
TM108_HUMAN
|
||||||
| NC score | 0.025073 (rank : 99) | θ value | 2.36792 (rank : 98) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6UXF1, Q9BQH1, Q9BW81, Q9C0H3 | Gene names | TMEM108, KIAA1690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
PLCB1_MOUSE
|
||||||
| NC score | 0.024984 (rank : 100) | θ value | 1.81305 (rank : 95) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z1B3, Q62075, Q8K5A5, Q8K5A6, Q9Z0E5, Q9Z2T5 | Gene names | Plcb1, Plcb | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
ARRS_HUMAN
|
||||||
| NC score | 0.023851 (rank : 101) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10523, Q53SV3, Q99858 | Gene names | SAG | |||
|
Domain Architecture |

|
|||||
| Description | S-arrestin (Retinal S-antigen) (48 kDa protein) (S-AG) (Rod photoreceptor arrestin). | |||||
|
PLCB1_HUMAN
|
||||||
| NC score | 0.023766 (rank : 102) | θ value | 1.38821 (rank : 90) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NQ66, O60325, Q5TFF7, Q5TGC9, Q8IV93, Q9BQW2, Q9H4H2, Q9H8H5, Q9NQ65, Q9NQH9, Q9NTH4, Q9UJP6, Q9UM26 | Gene names | PLCB1, KIAA0581 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
P3H3_HUMAN
|
||||||
| NC score | 0.023744 (rank : 103) | θ value | 0.813845 (rank : 80) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IVL6, Q13512, Q15740, Q66K32, Q6NX61, Q7L2T1 | Gene names | LEPREL2, P3H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B). | |||||
|
CCD62_HUMAN
|
||||||
| NC score | 0.023229 (rank : 104) | θ value | 1.81305 (rank : 92) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6P9F0, Q6ZVF2, Q86VJ0, Q9BYZ5 | Gene names | CCDC62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 60 (Aaa-protein) (Protein TSP- NY). | |||||
|
LRC45_HUMAN
|
||||||
| NC score | 0.022449 (rank : 105) | θ value | 0.365318 (rank : 70) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
DYH5_MOUSE
|
||||||
| NC score | 0.021898 (rank : 106) | θ value | 0.62314 (rank : 74) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
HPS5_HUMAN
|
||||||
| NC score | 0.021531 (rank : 107) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPZ3, O95942, Q8N4U0 | Gene names | HPS5, AIBP63, KIAA1017 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 5 protein (Alpha-integrin-binding protein 63) (Ruby-eye protein 2 homolog) (Ru2). | |||||
|
P5CS_HUMAN
|
||||||
| NC score | 0.021351 (rank : 108) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54886, O95952, Q9UM72 | Gene names | ALDH18A1, P5CS, PYCS | |||
|
Domain Architecture |

|
|||||
| Description | Delta 1-pyrroline-5-carboxylate synthetase (P5CS) (Aldehyde dehydrogenase 18 family member A1) [Includes: Glutamate 5-kinase (EC 2.7.2.11) (Gamma-glutamyl kinase) (GK); Gamma-glutamyl phosphate reductase (GPR) (EC 1.2.1.41) (Glutamate-5-semialdehyde dehydrogenase) (Glutamyl-gamma-semialdehyde dehydrogenase)]. | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.020920 (rank : 109) | θ value | 0.365318 (rank : 69) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
MBD5_HUMAN
|
||||||
| NC score | 0.020878 (rank : 110) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
S35B2_HUMAN
|
||||||
| NC score | 0.019651 (rank : 111) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TB61, Q5T9W1, Q5T9W2, Q7Z2G3, Q8NBK6, Q96AR6 | Gene names | SLC35B2, PAPST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenosine 3'-phospho 5'-phosphosulfate transporter 1 (PAPS transporter 1) (Solute carrier family 35 member B2) (Putative MAPK-activating protein PM15) (Putative NF-kappa-B-activating protein 48). | |||||
|
CT026_HUMAN
|
||||||
| NC score | 0.019332 (rank : 112) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NHU2, Q5TE18, Q8N5R9, Q96M59, Q9BQL2, Q9H127, Q9H128, Q9NQH4, Q9UFV8, Q9Y4V7 | Gene names | C20orf26 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf26. | |||||
|
PK3CB_HUMAN
|
||||||
| NC score | 0.019056 (rank : 113) | θ value | 1.06291 (rank : 86) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42338 | Gene names | PIK3CB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
CCM2_MOUSE
|
||||||
| NC score | 0.018957 (rank : 114) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2Y9 | Gene names | Ccm2, Osm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Malcavernin (Cerebral cavernous malformations protein 2 homolog) (Osmosensing scaffold for MEKK3). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.018753 (rank : 115) | θ value | 0.813845 (rank : 79) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
RABE1_MOUSE
|
||||||
| NC score | 0.017896 (rank : 116) | θ value | 2.36792 (rank : 97) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
RCOR1_HUMAN
|
||||||
| NC score | 0.017623 (rank : 117) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKL0, Q15044, Q6P2I9, Q86VG5 | Gene names | RCOR1, KIAA0071, RCOR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR1_MOUSE
|
||||||
| NC score | 0.017437 (rank : 118) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CFE3, Q3V092, Q8BK28, Q8CHI2 | Gene names | Rcor1, D12Wsu95e, Kiaa0071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
SFRP5_HUMAN
|
||||||
| NC score | 0.017273 (rank : 119) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T4F7, O14780, Q86TH7 | Gene names | SFRP5, FRP1B, SARP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 5 precursor (sFRP-5) (Secreted apoptosis-related protein 3) (SARP-3) (Frizzled-related protein 1b) (FRP-1b). | |||||
|
CCM2_HUMAN
|
||||||
| NC score | 0.017187 (rank : 120) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BSQ5, Q71RE5, Q8TAT4 | Gene names | CCM2, C7orf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Malcavernin (Cerebral cavernous malformations 2 protein). | |||||
|
PXDC1_MOUSE
|
||||||
| NC score | 0.016641 (rank : 121) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91ZV7, Q8BM20, Q9CWV5 | Gene names | Plxdc1, Tem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 1 precursor (Tumor endothelial marker 7). | |||||
|
FOXP4_HUMAN
|
||||||
| NC score | 0.016191 (rank : 122) | θ value | 0.47712 (rank : 71) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IVH2, Q96E19 | Gene names | FOXP4, FKHLA | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P4 (Fork head-related protein-like A). | |||||
|
FA45B_HUMAN
|
||||||
| NC score | 0.016190 (rank : 123) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6NSW5, Q9NZ36 | Gene names | FAM45B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM45B. | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.016062 (rank : 124) | θ value | 0.62314 (rank : 73) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
PTRF_MOUSE
|
||||||
| NC score | 0.016049 (rank : 125) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O54724 | Gene names | Ptrf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polymerase I and transcript release factor. | |||||
|
TRAIP_HUMAN
|
||||||
| NC score | 0.015653 (rank : 126) | θ value | 4.03905 (rank : 118) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.015642 (rank : 127) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
PK3CB_MOUSE
|
||||||
| NC score | 0.015391 (rank : 128) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BTI9 | Gene names | Pik3cb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
CN037_MOUSE
|
||||||
| NC score | 0.014961 (rank : 129) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
FAS_MOUSE
|
||||||
| NC score | 0.014881 (rank : 130) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19096, Q6PB72, Q8C4Z0, Q9EQR0 | Gene names | Fasn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.014666 (rank : 131) | θ value | 1.06291 (rank : 83) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
ANR18_HUMAN
|
||||||
| NC score | 0.014644 (rank : 132) | θ value | 0.813845 (rank : 78) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1106 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IVF6, Q7Z468 | Gene names | ANKRD18A, KIAA2015 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 18A. | |||||
|
SFRP1_HUMAN
|
||||||
| NC score | 0.014486 (rank : 133) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N474, O00546, O14779 | Gene names | SFRP1, FRP, FRP1, SARP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 1 precursor (sFRP-1) (Frizzled- related protein 1) (FRP-1) (Secreted apoptosis-related protein 2) (SARP-2). | |||||
|
SFRP5_MOUSE
|
||||||
| NC score | 0.014161 (rank : 134) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WU66, Q8K269 | Gene names | Sfrp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 5 precursor (sFRP-5). | |||||
|
SPTN5_HUMAN
|
||||||
| NC score | 0.013786 (rank : 135) | θ value | 1.38821 (rank : 91) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.013580 (rank : 136) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
BCL7C_MOUSE
|
||||||
| NC score | 0.013572 (rank : 137) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08664, Q99LS2 | Gene names | Bcl7c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C (B-cell chronic lymphocytic leukemia/lymphoma 7C protein). | |||||
|
CRGB_HUMAN
|
||||||
| NC score | 0.013300 (rank : 138) | θ value | 1.81305 (rank : 93) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07316 | Gene names | CRYGB, CRYG2 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma crystallin B (Gamma crystallin 1-2). | |||||
|
TNR1B_MOUSE
|
||||||
| NC score | 0.013041 (rank : 139) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P25119, O88734, P97893 | Gene names | Tnfrsf1b, Tnfr-2, Tnfr2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor). | |||||
|
VWF_MOUSE
|
||||||
| NC score | 0.012966 (rank : 140) | θ value | 1.06291 (rank : 88) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
MLH1_MOUSE
|
||||||
| NC score | 0.012835 (rank : 141) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JK91, Q62454 | Gene names | Mlh1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh1 (MutL protein homolog 1). | |||||
|
K0859_HUMAN
|
||||||
| NC score | 0.012169 (rank : 142) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N6R0, O94940, Q5TGP9, Q5TGQ0, Q8N2P8, Q96J11, Q96SQ0, Q9Y2Z1, Q9Y3M6 | Gene names | KIAA0859 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0859. | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.012114 (rank : 143) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
LARGE_MOUSE
|
||||||
| NC score | 0.011970 (rank : 144) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1M7, Q497S9, Q6P7U2, Q80TW0 | Gene names | Large, Kiaa0609, Large1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycosyltransferase-like protein LARGE1 (EC 2.4.-.-) (Acetylglucosaminyltransferase-like 1A). | |||||
|
TACC3_HUMAN
|
||||||
| NC score | 0.011638 (rank : 145) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.011526 (rank : 146) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
PKD1_HUMAN
|
||||||
| NC score | 0.011053 (rank : 147) | θ value | 3.0926 (rank : 105) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
ANR38_MOUSE
|
||||||
| NC score | 0.011000 (rank : 148) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 512 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P9J5, Q8BV03 | Gene names | Ankrd38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 38. | |||||
|
GAS8_HUMAN
|
||||||
| NC score | 0.010888 (rank : 149) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
MBN3_HUMAN
|
||||||
| NC score | 0.009759 (rank : 150) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUK0, Q5JXN8, Q5JXN9, Q5JXP4, Q6UDQ1, Q8TAD9, Q8TAF4, Q9H0Z7, Q9UF37 | Gene names | MBNL3, CHCR, MBLX39, MBXL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein HCHCR). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.009670 (rank : 151) | θ value | 1.06291 (rank : 85) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
PAR14_MOUSE
|
||||||
| NC score | 0.009421 (rank : 152) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q2EMV9, Q571C6, Q8BN35, Q8VDT6 | Gene names | Parp14, Kiaa1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (Collaborator of STAT6) (CoaSt6). | |||||
|
IF122_HUMAN
|
||||||
| NC score | 0.009114 (rank : 153) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HBG6 | Gene names | IFT122, WDR10, WDR140 | |||
|
Domain Architecture |

|
|||||
| Description | Intraflagellar transport 122 homolog (WD repeat protein 10). | |||||
|
DNAI2_HUMAN
|
||||||
| NC score | 0.009060 (rank : 154) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9GZS0, Q9H179, Q9NT53 | Gene names | DNAI2 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein intermediate chain 2, axonemal (Axonemal dynein intermediate chain 2). | |||||
|
STX1A_MOUSE
|
||||||
| NC score | 0.008618 (rank : 155) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35526 | Gene names | Stx1a | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-1A (Neuron-specific antigen HPC-1). | |||||
|
STX1A_HUMAN
|
||||||
| NC score | 0.008460 (rank : 156) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16623, O15447, O15448, Q12936, Q9BPZ6 | Gene names | STX1A, STX1 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-1A (Neuron-specific antigen HPC-1). | |||||
|
TBD_HUMAN
|
||||||
| NC score | 0.008308 (rank : 157) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UJT1, Q9BWG9, Q9H7Z8 | Gene names | TUBD1, TUBD | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin delta chain (Delta tubulin). | |||||
|
ANR15_HUMAN
|
||||||
| NC score | 0.007646 (rank : 158) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14678, Q5W0W0, Q8IY65, Q8WX74 | Gene names | ANKRD15, KANK, KIAA0172 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 15 (Kidney ankyrin repeat- containing protein). | |||||
|
WD51B_MOUSE
|
||||||
| NC score | 0.007186 (rank : 159) | θ value | 2.36792 (rank : 101) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BHD1 | Gene names | Wdr51b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 51B. | |||||
|
BRCA1_HUMAN
|
||||||
| NC score | 0.006693 (rank : 160) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38398 | Gene names | BRCA1, RNF53 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein (RING finger protein 53). | |||||
|
WD51B_HUMAN
|
||||||
| NC score | 0.006631 (rank : 161) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TC44 | Gene names | WDR51B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 51B. | |||||
|
LRC19_HUMAN
|
||||||
| NC score | 0.006561 (rank : 162) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H756 | Gene names | LRRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 19 precursor. | |||||
|
K1C14_MOUSE
|
||||||
| NC score | 0.006409 (rank : 163) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61781, Q91VQ4 | Gene names | Krt14, Krt1-14 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 14 (Cytokeratin-14) (CK-14) (Keratin-14) (K14). | |||||
|
MTMR9_MOUSE
|
||||||
| NC score | 0.005846 (rank : 164) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2D0, Q80XL4 | Gene names | Mtmr9, Mtmr3 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 9. | |||||
|
RPGF2_HUMAN
|
||||||
| NC score | 0.005621 (rank : 165) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4G8 | Gene names | RAPGEF2, KIAA0313, NRAPGEP, PDZGEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rap guanine nucleotide exchange factor 2 (Neural RAP guanine nucleotide exchange protein) (nRap GEP) (PDZ domain-containing guanine nucleotide exchange factor 1) (PDZ-GEF1) (RA-GEF). | |||||
|
GP176_MOUSE
|
||||||
| NC score | 0.005556 (rank : 166) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80WT4, Q80UC4 | Gene names | Gpr176, Agr9, Gm1012 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 176 (G-protein coupled receptor AGR9). | |||||
|
BRD3_HUMAN
|
||||||
| NC score | 0.005543 (rank : 167) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15059, Q5T1R7, Q8N5M3, Q92645 | Gene names | BRD3, KIAA0043, RING3L | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (RING3-like protein). | |||||
|
ABCAD_MOUSE
|
||||||
| NC score | 0.005167 (rank : 168) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SSE9, Q80T20, Q8BHZ2, Q8CB91 | Gene names | Abca13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
K1C14_HUMAN
|
||||||
| NC score | 0.004553 (rank : 169) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P02533, Q14715, Q53XY3, Q9BUE3, Q9UBN2, Q9UBN3, Q9UCY4 | Gene names | KRT14 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 14 (Cytokeratin-14) (CK-14) (Keratin-14) (K14). | |||||
|
CAC1I_HUMAN
|
||||||
| NC score | 0.003399 (rank : 170) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
PO6F1_MOUSE
|
||||||
| NC score | 0.002710 (rank : 171) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
MPDZ_HUMAN
|
||||||
| NC score | 0.002587 (rank : 172) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75970, O43798, Q5CZ80, Q5JTX3, Q5JTX6, Q5JTX7, Q5JUC3, Q5JUC4, Q5VZ62, Q8N790 | Gene names | MPDZ, MUPP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ- domain protein 1). | |||||
|
EMR1_MOUSE
|
||||||
| NC score | 0.000772 (rank : 173) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61549 | Gene names | Emr1, Gpf480 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 1 precursor (Cell surface glycoprotein EMR1) (EMR1 hormone receptor) (Cell surface glycoprotein F4/80). | |||||
|
EMR1_HUMAN
|
||||||
| NC score | 0.000743 (rank : 174) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14246, Q8NGA7 | Gene names | EMR1, TM7LN3 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 1 precursor (Cell surface glycoprotein EMR1) (EMR1 hormone receptor). | |||||
|
OX1R_HUMAN
|
||||||
| NC score | -0.000020 (rank : 175) | θ value | 1.81305 (rank : 94) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 909 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43613 | Gene names | HCRTR1 | |||
|
Domain Architecture |

|
|||||
| Description | Orexin receptor type 1 (Ox1r) (Hypocretin receptor type 1). | |||||
|
GAK_HUMAN
|
||||||
| NC score | -0.002963 (rank : 176) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
O5BF1_HUMAN
|
||||||
| NC score | -0.004585 (rank : 177) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NHC7 | Gene names | OR5BF1 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 5BF1. | |||||